5AHI
 
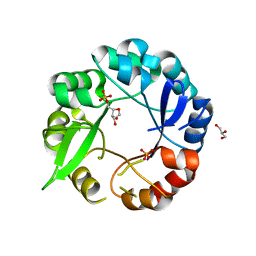 | | Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanism of Hisa from Salmonella Enterica
To be Published
|
|
6PM7
 
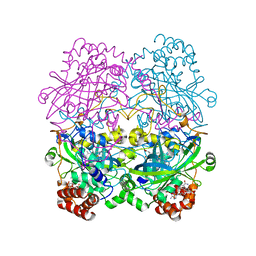 | |
6B15
 
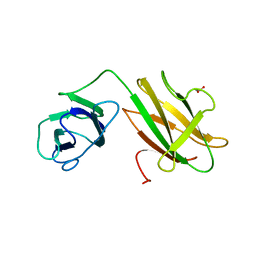 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K | | Descriptor: | 1,2-ETHANEDIOL, Amy13K | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-16 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
7TQR
 
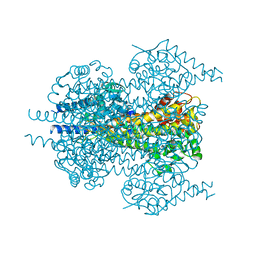 | |
6N0M
 
 | |
5ADB
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-((4-Chloro-3-((methylamino)methyl)phenoxy)methyl) quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
8DB1
 
 | | Crystal structure of native DMATS1 prenyltransferase | | Descriptor: | Dimethylallyltryptophan synthase 1, TRYPTOPHAN | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
6PMY
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(2-aminoethyl)phenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7P4P
 
 | |
6PN9
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(thiazol-5-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(1,3-thiazol-5-yl)methoxy]phenyl}-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5A2O
 
 | |
7U69
 
 | |
5A36
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
5ACV
 
 | | VIM-2-OX, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
7U3M
 
 | |
7U6A
 
 | |
6PMP
 
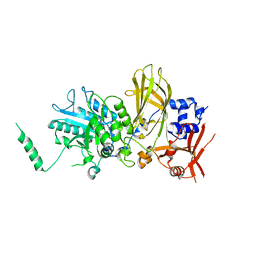 | |
5AED
 
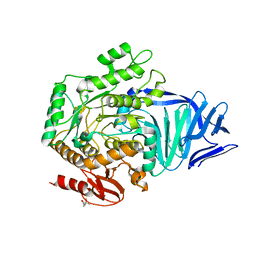 | | A bacterial protein structure in glycoside hydrolase family 31 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-GLUCOSIDASE YIHQ, CALCIUM ION | | Authors: | Jin, Y, Speciale, G, Davies, G.J, Williams, S.J, Goddard-Borger, E.D. | | Deposit date: | 2015-08-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Yihq is a Sulfoquinovosidase that Cleaves Sulfoquinovosyl Diacylglyceride Sulfolipids.
Nat.Chem.Biol., 12, 2016
|
|
7P6V
 
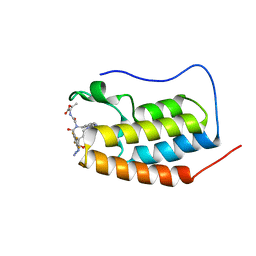 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH compound 3ag | | Descriptor: | Bromodomain-containing protein 4, ethyl 2-(2-(4-azido-N-((2-(1,5-dimethyl-6-oxo-1,6-dihydropyridin-3-yl)-1-((tetrahydro-2H-pyran-4-yl)methyl)-1H-benzo[d]imidazol-6-yl)methyl)-2,3,5,6-tetrafluorobenzamido)acetamido)acetate | | Authors: | Chung, C. | | Deposit date: | 2021-07-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | One-Step Synthesis of Photoaffinity Probes for Live-Cell MS-Based Proteomics.
Chemistry, 27, 2021
|
|
8DYH
 
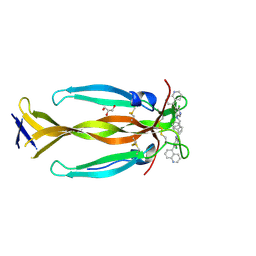 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
7TLX
 
 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
7TOK
 
 | | Crystal structure of the CBM domain of carbohydrate esterase FjoAcXE | | Descriptor: | Acetylxylan esterase I | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
6PO5
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclobutylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclobutylmethoxy)phenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6VES
 
 | | Human insulin analog: [GluB10,HisA8,ArgA9]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mini-Ins: A Minimal, Bioactive Insulin Analog with Alternative Binding Modes
To Be Published
|
|
6PPV
 
 | |
