6XYI
 
 | | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3) | | Descriptor: | AMS9.3.1 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3)
TO BE PUBLISHED
|
|
4OS7
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK607 (bicyclic) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS5
 
 | |
4OS1
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK601 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS2
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK602 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
1V6P
 
 | | Crystal structure of Cobrotoxin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Cobrotoxin, ... | | Authors: | Lou, X, Tu, X, Wang, J, Teng, M, Niu, L, Liu, Q, Huang, Q, Hao, Q. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
4OS6
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK604 (bicyclic 2) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS4
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK603 (bicyclic 1) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
1VB0
 
 | | Atomic resolution structure of atratoxin-b, one short-chain neurotoxin from Naja atra | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Liu, Q, Teng, M, Niu, L, Huang, Q, Hao, Q. | | Deposit date: | 2004-02-20 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
2AJW
 
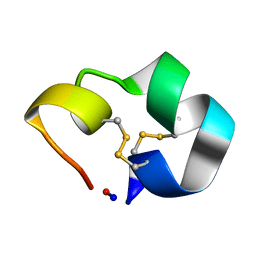 | | Structure of the cyclic conotoxin MII-6 | | Descriptor: | Alpha-conotoxin MII | | Authors: | Clark, R.J, Fischer, H, Dempster, L, Daly, N.L, Rosengren, K.J, Nevin, S.T, Meunier, F.A, Adams, D.J, Craik, D.J. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the {alpha}-conotoxin MII.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1COE
 
 | |
2AK0
 
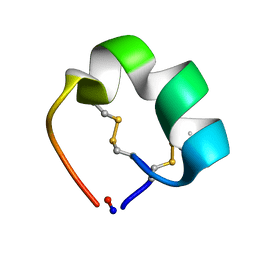 | | Structure of cyclic conotoxin MII-7 | | Descriptor: | Alpha-conotoxin MII | | Authors: | Clark, R.J, Fischer, H, Dempster, L, Daly, N.L, Rosengren, K.J, Nevin, S.T, Meunier, F.A, Adams, D.J, Craik, D.J. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the {alpha}-conotoxin MII.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1COD
 
 | |
1NOR
 
 | | TWO-DIMENSIONAL 1H-NMR STUDY OF THE SPATIAL STRUCTURE OF NEUROTOXIN II FROM NAJA OXIANA | | Descriptor: | NEUROTOXIN II | | Authors: | Golovanov, A.P, Utkin, Y.N, Lomize, A.L, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 1993-04-05 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H-NMR study of the spatial structure of neurotoxin II from Naja naja oxiana.
Eur.J.Biochem., 213, 1993
|
|
2NS3
 
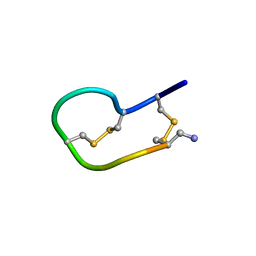 | | Solution structure of ribbon BuIA | | Descriptor: | Alpha-conotoxin BuIA | | Authors: | Jin, A.H, Brandstaetter, H, Nevin, S.T, Tan, C.C, Clark, R.J, Adams, D.J, Alewood, P.F, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-11-03 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of alpha-conotoxin BuIA: influences of disulfide connectivity on structural dynamics
Bmc Struct.Biol., 7, 2007
|
|
1NTN
 
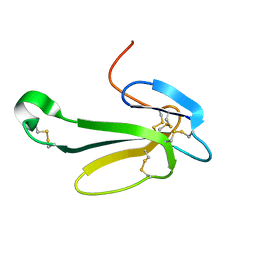 | | THE CRYSTAL STRUCTURE OF NEUROTOXIN-I FROM NAJA NAJA OXIANA AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | NEUROTOXIN I | | Authors: | Mikhailov, A.M, Nickitenko, A.V, Vainshtein, B.K, Betzel, C, Wilson, K. | | Deposit date: | 1994-09-26 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structure of neurotoxin-1 from Naja naja oxiana venom at 1.9 A resolution.
Febs Lett., 320, 1993
|
|
1NTX
 
 | |
1QMW
 
 | | Solution structure of alpha-conotoxin SI | | Descriptor: | ALPHA-CONOTOXIN SI | | Authors: | Benie, A.J, Whitford, D, Hargittai, B, Barany, G, Janes, R.W. | | Deposit date: | 1999-10-08 | | Release date: | 2000-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Alpha-Conotoxin Si
FEBS Lett., 476, 2000
|
|
1ONJ
 
 | | Crystal structure of Atratoxin-b from Chinese cobra venom of Naja atra | | Descriptor: | Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Tu, X, Pan, G, Xu, C, Fan, R, Lu, W, Deng, W, Rao, P, Teng, M, Niu, L. | | Deposit date: | 2003-02-28 | | Release date: | 2004-02-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Purification, N-terminal sequencing, crystallization and preliminary structural determination of atratoxin-b, a short-chain alpha-neurotoxin from Naja atra venom.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1G6M
 
 | |
1IQ9
 
 | |
1JGK
 
 | |
2MJ4
 
 | |
7UI6
 
 | |
7UI7
 
 | |
