7YX1
 
 | |
8S5Y
 
 | | High pH (8.0) as-isolated MSOX movie series dataset 40 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [14 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8ON4
 
 | |
7YP3
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with elaiophylin | | Descriptor: | ACETATE ION, Elaiophylin, GLYCEROL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7N0Z
 
 | |
8S64
 
 | | Low pH (5.5) as-isolated MSOX movie series dataset 10 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [5.7 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8OND
 
 | |
5U99
 
 | | Transition state analysis of adenosine triphosphate phosphoribosyltransferase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Moggre, G.-J, Poulin, M.B, Tyler, P.C, Schramm, V.L, Parker, E.J. | | Deposit date: | 2016-12-15 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transition State Analysis of Adenosine Triphosphate Phosphoribosyltransferase.
ACS Chem. Biol., 12, 2017
|
|
8OK0
 
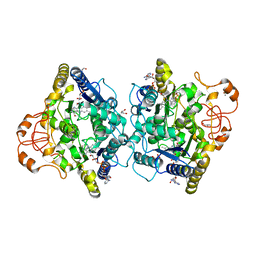 | | Crystal structure of human NQO1 in complex with the inhibitor PMSF | | Descriptor: | ACETYL GROUP, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A. | | Deposit date: | 2023-03-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dynamics at the active site of the cancer-associated flavoenzyme NQO1 probed by chemical modification with PMSF.
Febs Lett., 597, 2023
|
|
7NAA
 
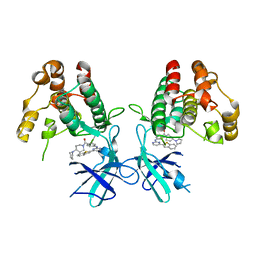 | | Crystal structure of Mycobacterium tuberculosis H37Rv PknF kinase domain | | Descriptor: | (4-{[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]amino}phenyl)[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone, Non-specific serine/threonine protein kinase | | Authors: | Oliveira, A.A, Cabarca, S, dos Reis, C.V, Takarada, J.E, Counago, R.M, Balan, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Mycobacterium tuberculosis c PknF and conformational changes induced in forkhead-associated regulatory domains.
Curr Res Struct Biol, 3, 2021
|
|
5H4F
 
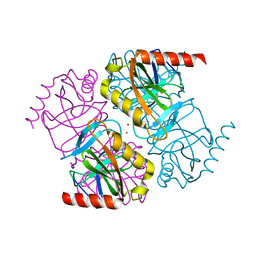 | | Structure of inorganic pyrophosphatase crystallised as a contaminant | | Descriptor: | ZINC ION, inorganic pyrophosphatase | | Authors: | Chaudhary, S, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
7PSY
 
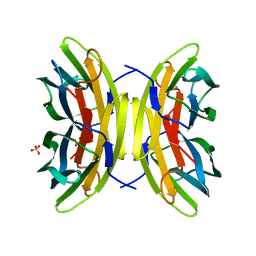 | | X-ray crystal structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
7VIV
 
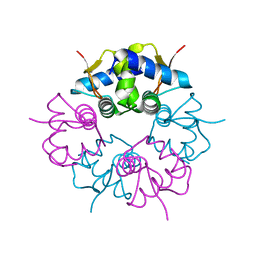 | |
8S8C
 
 | | Structure of Kras in complex with inhibitor MK-1084 | | Descriptor: | (5aSa,17aRa)- 20-Chloro-2-[(2S,5R)-2,5-dimethyl-4-(prop-2-enoyl)piperazin-1-yl]-14,17-difluoro-6-(propan-2-yl)-11,12-dihydro-4H-1,18-(ethanediylidene)pyrido[4,3-e]pyrimido[1,6-g][1,4,7,9]benzodioxadiazacyclododecin-4-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Day, P.J, Cleasby, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-1084: An Orally Bioavailable and Low-Dose KRAS G12C Inhibitor.
J.Med.Chem., 67, 2024
|
|
5CE6
 
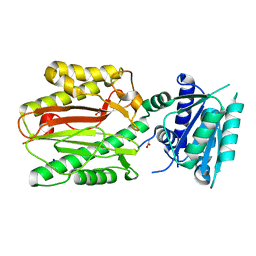 | | N-terminal domain of FACT complex subunit SPT16 from Cicer arietinum (chickpea) | | Descriptor: | ACETATE ION, FACT-Spt16, POTASSIUM ION, ... | | Authors: | Are, V.N, Ghosh, B, Kumar, A, Makde, R. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum.
Int.J.Biol.Macromol., 88, 2016
|
|
8P32
 
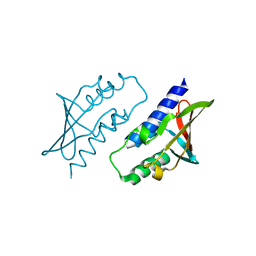 | | BB0238 from Borrelia burgdorferi, Se-Met data for Leu240Met mutant | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
8RFX
 
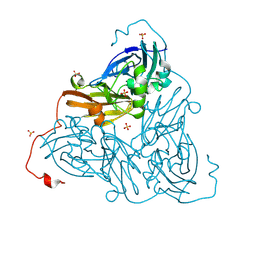 | | High pH (8.0) as-isolated MSOX movie series dataset 5 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [2.05 MGy] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7VBO
 
 | | Alginate binding domain CBM | | Descriptor: | Alginate lyase, CALCIUM ION, SULFATE ION | | Authors: | Ji, S.Q, She, Q. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification and structural analysis of a carbohydrate-binding module specific to alginate, a representative of a new family, CBM96.
J.Biol.Chem., 299, 2023
|
|
8RSV
 
 | |
8RFL
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 1 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [0.61 MGy] - nitrite (start) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8UC5
 
 | |
8UC4
 
 | |
8RGC
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 10 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [6.8 MGy] | | Descriptor: | CARBON DIOXIDE, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7N09
 
 | |
5CQ6
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2,6-Pyridinedicarboxylic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2,6-Pyridinedicarboxylic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
