7MI5
 
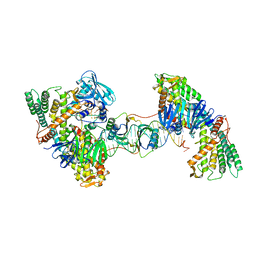 | | Asymmetrical PAM-Non PAM prespacer bound Cas4/Cas1/Cas2 complex | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (26-MER), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
2EXF
 
 | |
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
4Z2C
 
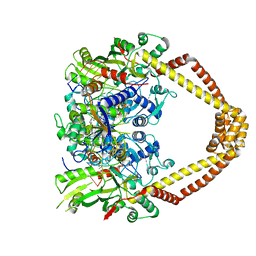 | | Quinolone(Moxifloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
4Z2D
 
 | | Quinolone(Levofloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
4MDF
 
 | | Structure of bacterial polynucleotide kinase Michaelis complex bound to GTP and DNA | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
1WRT
 
 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1WRS
 
 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
8C8J
 
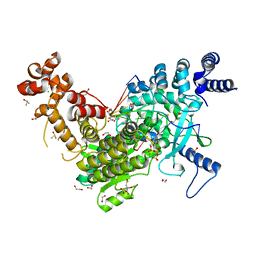 | | Long Interspersed Nuclear Element 1 (LINE-1) reverse transcriptase ternary complex with hybrid duplex and dTTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Walpole, T.B, Baldwin, E. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
2C6R
 
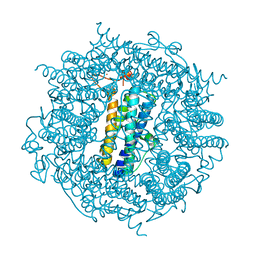 | | FE-SOAKED CRYSTAL STRUCTURE OF THE DPS92 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | CHLORIDE ION, DNA-BINDING STRESS RESPONSE PROTEIN, DPS FAMILY, ... | | Authors: | Cuypers, M.G, Romao, C.V, Mitchell, E, Mcsweeney, S. | | Deposit date: | 2005-11-11 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Dps2 from Deinococcus Radiodurans Reveals an Unusual Pore Profile with a Non-Specific Metal Binding Site.
J.Mol.Biol., 371, 2007
|
|
6V2K
 
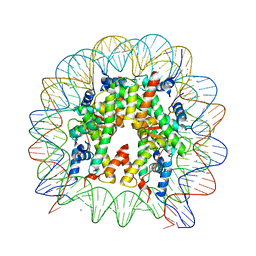 | | The nucleosome structure after H2A-H2B exchange | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Arimura, Y, Hirano, R, Kurumizaka, H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome.
Commun Biol, 4, 2021
|
|
7N10
 
 | |
2ZTE
 
 | | MtRuvA Form IV | | Descriptor: | Holliday junction ATP-dependent DNA helicase ruvA | | Authors: | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and modelling studies on Mycobacterium tuberculosis RuvA Additional role of RuvB-binding domain and inter species variability
Biochim.Biophys.Acta, 1794, 2009
|
|
6YSZ
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly | | Descriptor: | Internal virion protein gp15 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YT5
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15-gp16 core complex intermediate assembly | | Descriptor: | Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2ZTC
 
 | | MtRuvA Form II | | Descriptor: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | Authors: | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and modelling studies on Mycobacterium tuberculosis RuvA Additional role of RuvB-binding domain and inter species variability
Biochim.Biophys.Acta, 1794, 2009
|
|
2ZTD
 
 | | MtRuvA Form III | | Descriptor: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | Authors: | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and modelling studies on Mycobacterium tuberculosis RuvA Additional role of RuvB-binding domain and inter species variability
Biochim.Biophys.Acta, 1794, 2009
|
|
1AK0
 
 | | P1 NUCLEASE IN COMPLEX WITH A SUBSTRATE ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-(DITHIO)PHOSPHATE, ... | | Authors: | Romier, C, Suck, D. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs.
Proteins, 32, 1998
|
|
1ENJ
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
6B8A
 
 | | Crystal structure of MvfR ligand binding domain in complex with M64 | | Descriptor: | 2-[(5-nitro-1H-benzimidazol-2-yl)sulfanyl]-N-(4-phenoxyphenyl)acetamide, COBALT HEXAMMINE(III), DNA-binding transcriptional regulator | | Authors: | Kitao, T, Steinbacher, S, Maskos, K, Blaesse, M, Rahme, L.G. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Insights into Function and Competitive Inhibition ofPseudomonas aeruginosaMultiple Virulence Factor Regulator.
MBio, 9, 2018
|
|
4QH0
 
 | | Crystal structure of NucA from Streptococcus agalactiae with magnesium ion bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Moon, A.F, Gaudu, P, Pedersen, L.C. | | Deposit date: | 2014-05-26 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7EI5
 
 | |
6IYA
 
 | |
1DSD
 
 | |
1DSC
 
 | |
