4V40
 
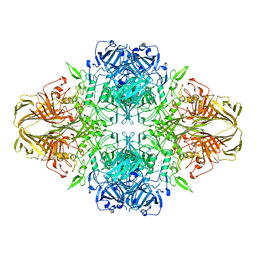 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
4UXJ
 
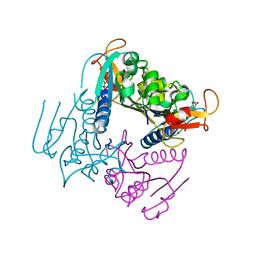 | | Leishmania major Thymidine Kinase in complex with dTTP | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
2REK
 
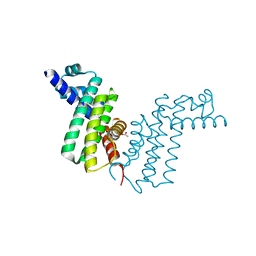 | | Crystal structure of tetR-family transcriptional regulator | | Descriptor: | ACETATE ION, Putative tetR-family transcriptional regulator | | Authors: | Dong, A, Xu, X, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of tetR-family transcriptional regulator.
To be Published
|
|
4WIZ
 
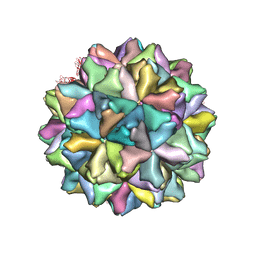 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4WGF
 
 | | YcaC from Pseudomonas aeruginosa with hexane-2,5-diol and covalent acrylamide | | Descriptor: | (2R,5R)-hexane-2,5-diol, CHLORIDE ION, PROPIONAMIDE, ... | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34020686 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
2RF7
 
 | | Crystal structure of the escherichia coli nrfa mutant Q263E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
6FIT
 
 | | FHIT-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE MONOTUNGSTATE, FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
721P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
6EST
 
 | |
2VUU
 
 | | Crystal structure of NADP-bound NmrA-AreA zinc finger complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
3JZP
 
 | |
7AT1
 
 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
7AHL
 
 | | ALPHA-HEMOLYSIN FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | ALPHA-HEMOLYSIN | | Authors: | Song, L, Hobaugh, M, Shustak, C, Cheley, S, Bayley, H, Gouaux, J.E. | | Deposit date: | 1996-12-02 | | Release date: | 1998-01-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore.
Science, 274, 1996
|
|
2NW4
 
 | | Crystal Structure of the Rat Androgen Receptor Ligand Binding Domain Complex with BMS-564929 | | Descriptor: | 2-CHLORO-4-[(7R,7AS)-7-HYDROXY-1,3-DIOXOTETRAHYDRO-1H-PYRROLO[1,2-C]IMIDAZOL-2(3H)-YL]-3-METHYLBENZONITRILE, Androgen receptor | | Authors: | Ostrowski, J, Kuhns, J.E, Lupisella, J.A, Manfredi, M.C, Beehler, B.C, Krystek, S.R, Bi, Y, Sun, C, Seethala, R, Golla, R, Sleph, P.G, Fura, A, An, Y, Kish, K.F, Sack, J.S, Mookhtiar, K.A, Grover, G.J, Hamann, L.G. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pharmacological and x-ray structural characterization of a novel selective androgen receptor modulator: potent hyperanabolic stimulation of skeletal muscle with hypostimulation of prostate in rats.
Endocrinology, 148, 2007
|
|
3JZO
 
 | | Human MDMX liganded with a 12mer peptide (pDI) | | Descriptor: | POTASSIUM ION, Protein Mdm4, pDI peptide (12mer) | | Authors: | Schonbrunn, E, Phan, J. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX.
J.Biol.Chem., 285, 2010
|
|
2VUT
 
 | | Crystal structure of NAD-bound NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
6GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
7B72
 
 | | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(RGT)P*G)-3') | | Authors: | Lomzov, A.A, Pyshnyi, D.V, Shernuykov, A.V, Apukhtina, V.S. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2023-08-23 | | Method: | SOLUTION NMR | | Cite: | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer
To Be Published
|
|
7B7O
 
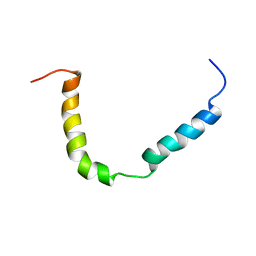 | | Solution structure of A. thaliana core TatA in DHPC micelles | | Descriptor: | Sec-independent protein translocase protein TATA, chloroplastic | | Authors: | Pettersson, P, Ye, W, Jakob, M, Tannert, F, Klosgen, R.B, Maler, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of plant TatA in micelles and lipid bilayers studied by solution NMR.
FEBS J, 285, 2018
|
|
7B71
 
 | | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(SGT)P*G)-3') | | Authors: | Lomzov, A.A, Shernuykov, A.V, Apukhtina, V.S, Pyshnyi, D.V. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer
To Be Published
|
|
2NXH
 
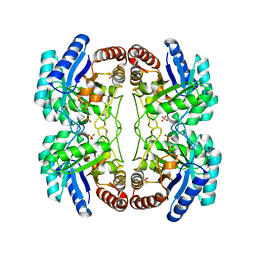 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase. | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHATE ION, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
7CGT
 
 | |
2NX3
 
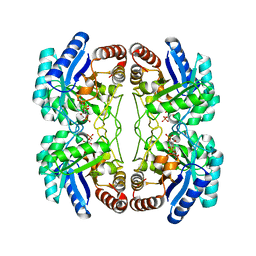 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase | | Descriptor: | (2R,4R,5R,6R,7R)-2,4,5,6,7-PENTAHYDROXY-2,8-BIS(PHOSPHONOOXY)OCTANOIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, ... | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
2NXI
 
 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase. | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHATE ION, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-17 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic changes along an engineered path from metallo to nonmetallo 3-deoxy-D-manno-octulosonate 8-phosphate synthases
Biochemistry, 46, 2007
|
|
7AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
