8UUY
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with inhibitor Jun12129 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
6MSW
 
 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans, K184L mutant | | Descriptor: | Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.169 Å) | | Cite: | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
8OQ9
 
 | | Crystal structure of the titin domain Fn3-56 | | Descriptor: | CHLORIDE ION, Titin, ZINC ION | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
7NG3
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
6MT9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
8RV5
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 1 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
7VJY
 
 | |
8ORL
 
 | |
7VK3
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.10 A resolution-2 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Guven, O. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
8UFR
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-methyl-7-(4-methyl-2,3,4,5-tetrahydrobenzo[f][1,4]oxazepin-7-yl)quinolin-2-amine dihydrochloride | | Descriptor: | (7M)-4-methyl-7-(4-methyl-2,3,4,5-tetrahydro-1,4-benzoxazepin-7-yl)quinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
8RV7
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 4 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-oxidanylprop-1-ynyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
7VK1
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 1.93 A resolution-5 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Ertem, B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7NF5
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7VK6
 
 | |
5CEJ
 
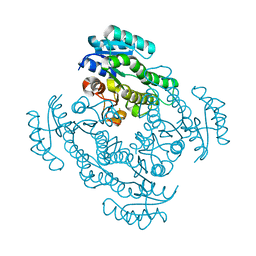 | |
8OTY
 
 | | Crystal structure of the titin domain Fn3-90 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
7NMQ
 
 | | Bacillus cereus HblL1 toxin component | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hemolysin BL lytic component L1 | | Authors: | Rizkallah, P.J, Berry, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The Crystal Structure of Bacillus cereus HblL 1 .
Toxins, 13, 2021
|
|
5CEP
 
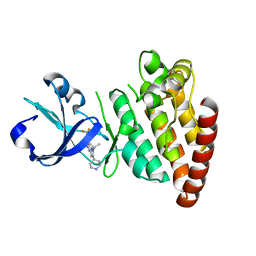 | |
5TQU
 
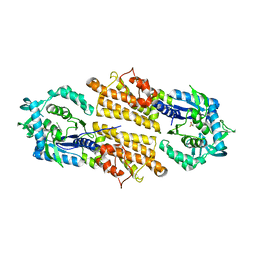 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor | | Descriptor: | 2-[2-[[3-(1~{H}-benzimidazol-2-ylamino)propylamino]methyl]-4,6-bis(chloranyl)indol-1-yl]ethanol, GLYCEROL, METHIONINE, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Cells to Mice to Target: Characterization of NEU-1053 (SB-443342) and Its Analogues for Treatment of Human African Trypanosomiasis.
ACS Infect Dis, 3, 2017
|
|
8UDC
 
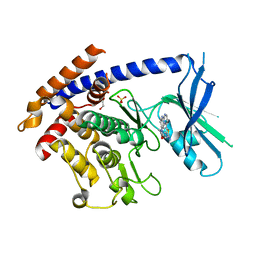 | | Crystal structure of TcPINK1 in complex with CYC116 | | Descriptor: | (4P)-4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-[4-(morpholin-4-yl)phenyl]pyrimidin-2-amine, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification and structural characterization of small molecule inhibitors of PINK1.
Sci Rep, 14, 2024
|
|
5H2R
 
 | | Crystal structure of T brucei phosphodiesterase B2 bound to compound 15b | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-(2-hydroxyethyloxy)-3-[2-(2-oxidanylideneimidazolidin-1-yl)ethoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, MAGNESIUM ION, Phosphodiesterase, ... | | Authors: | Noble, C.G. | | Deposit date: | 2016-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trypanosomal Phosphodiesterase B1 and B2 as a Potential Therapy for Human African Trypanosomiasis
To Be Published
|
|
6N4O
 
 | |
8RVA
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 7 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8OHA
 
 | | Crystal structure of Leptospira interrogans GAPDH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Navas-Yuste, S, de la Paz, K, Querol-Garcia, J, Gomez-Quevedo, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity.
Front Immunol, 14, 2023
|
|
8OIY
 
 | |
