4KTK
 
 | |
4K0A
 
 | |
1EU5
 
 | | STRUCTURE OF E. COLI DUTPASE AT 1.45 A | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, GLYCEROL | | Authors: | Gonzalez, A, Larsson, G, Persson, R, Cedergren-Zeppezauer, E. | | Deposit date: | 2000-04-13 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EOH
 
 | | GLUTATHIONE TRANSFERASE P1-1 | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | Deposit date: | 2000-03-22 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
1EWW
 
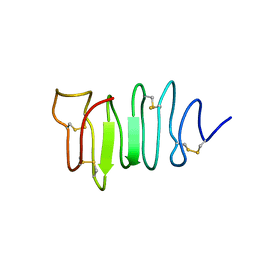 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
1EQ8
 
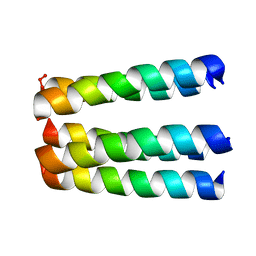 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, HYDROXIDE ION | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-26 | | Last modified: | 2022-02-16 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
4K2S
 
 | | Crystal structure of the mutant P317A of d-mannonate dehydratase from chromohalobacter salexigens complexed with mg and d-gluconate | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-mannonate dehydratase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of the mutant P317A of d-mannonate dehydratase from chromohalobacter salexigens complexed with mg and d-gluconate
To be Published
|
|
5I4H
 
 | |
4K7L
 
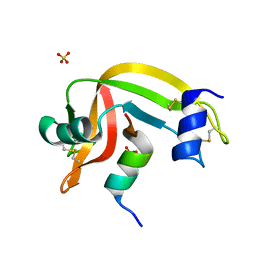 | | Crystal structure of RNase S variant (K7C/Q11C) | | Descriptor: | Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Straeter, N. | | Deposit date: | 2013-04-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of RNase S with a [Hg(Cys2)] metal center in the S-peptide as a template for structure-based design of artificial metalloenzymes using peptide-protein complementation
To be Published
|
|
1EVF
 
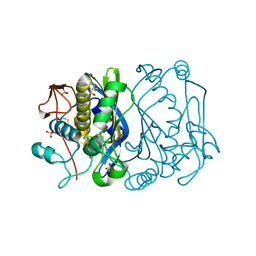 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EVR
 
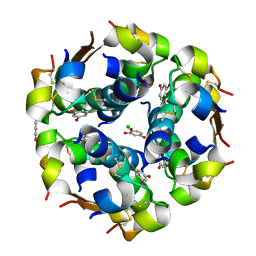 | | The structure of the resorcinol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, RESORCINOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-20 | | Release date: | 2000-12-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4KR2
 
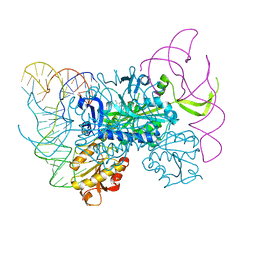 | | Glycyl-tRNA synthetase in complex with tRNA-Gly | | Descriptor: | ADENOSINE MONOPHOSPHATE, Gly-tRNA-CCC, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-16 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Cocrystal Structures of Glycyl-tRNA Synthetase in Complex with tRNA Suggest Multiple Conformational States in Glycylation
J.Biol.Chem., 289, 2014
|
|
5IRV
 
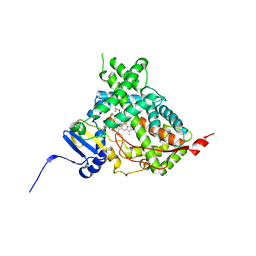 | | Human cytochrome P450 17A1 bound to inhibitor VT-464 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase, VT-464 | | Authors: | Scott, E.E, Petrunak, E.M. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural and Functional Evaluation of Clinically Relevant Inhibitors of Steroidogenic Cytochrome P450 17A1.
Drug Metab. Dispos., 45, 2017
|
|
4KR6
 
 | |
1EK2
 
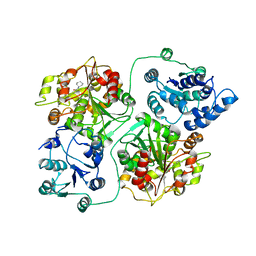 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CDU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-DECYLUREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
4K2A
 
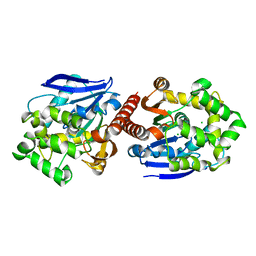 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | Deposit date: | 2013-04-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1T1C
 
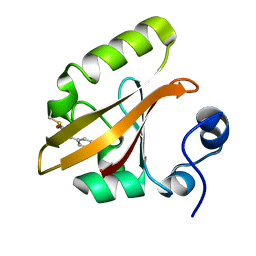 | | Late intermediate IL3 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1IHN
 
 | |
4K4H
 
 | |
1IK3
 
 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 13(S)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID | | Descriptor: | (TRANS-12,13-EPOXY)-11-HYDROXY-9(Z)-OCTADECENOIC ACID, (TRANS-12,13-EPOXY)-9-HYDROXY-10(E)-OCTADECENOIC ACID, 13(R)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID, ... | | Authors: | Skrzypczak-Jankun, E, Funk Jr, M.O. | | Deposit date: | 2001-05-01 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a purple lipoxygenase.
J.Am.Chem.Soc., 123, 2001
|
|
4K9D
 
 | |
4K79
 
 | | Recognition of the Thomsen-Friedenreich Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
4K9J
 
 | | Structure of Re(CO)3(4,7-dimethyl-phen)(Thr126His)(Lys122Trp)(His83Glu)(Trp48Phe)(Tyr72Phe)(Tyr108Phe)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Williamson, H.R, Blanco-Rodriguez, A.M, Sokolova, L, Nikolovski, P, Kaiser, J.T, Towrie, M, Clark, I.P, Vlcek Jr, A, Winkler, J.R, Gray, H.B. | | Deposit date: | 2013-04-20 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan-accelerated electron flow across a protein-protein interface.
J.Am.Chem.Soc., 135, 2013
|
|
1SY3
 
 | | 1.00 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1IN6
 
 | | THERMOTOGA MARITIMA RUVB K64R MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
