4F7G
 
 | |
1QAB
 
 | |
1QO6
 
 | |
1QHN
 
 | | CHLORAMPHENICOL PHOSPHOTRANSFERASE FROM STREPTOMYCES VENEZUELAE | | Descriptor: | CHLORAMPHENICOL PHOSPHOTRANSFERASE, SULFATE ION | | Authors: | Izard, T. | | Deposit date: | 1999-05-23 | | Release date: | 2000-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structures of Chloramphenicol Phosphotransferase Reveal a Novel Inactivation Mechanism
Embo J., 19, 2000
|
|
3PUO
 
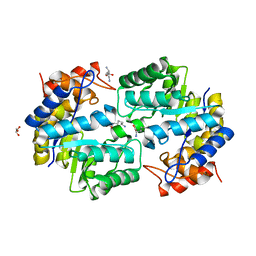 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
4M9Z
 
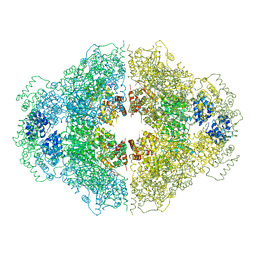 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
3R3J
 
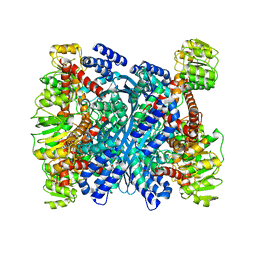 | | Kinetic and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2 | | Descriptor: | Glutamate dehydrogenase | | Authors: | Zocher, K, Fritz-Wolf, K, Kehr, S, Rahlfs, S, Becker, K. | | Deposit date: | 2011-03-16 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2.
Mol.Biochem.Parasitol., 183, 2012
|
|
4MDN
 
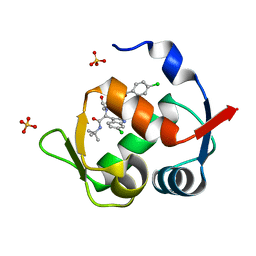 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
2VGX
 
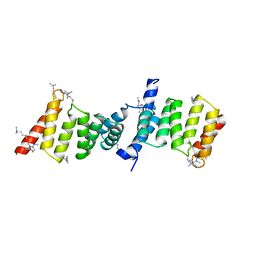 | | Structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
1Q7F
 
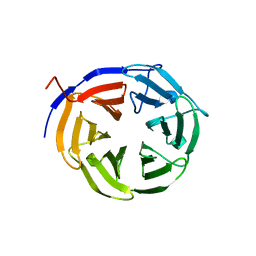 | |
4JNH
 
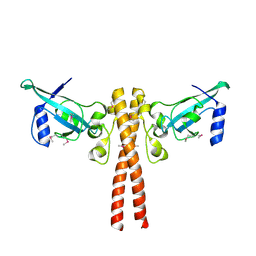 | |
4D6X
 
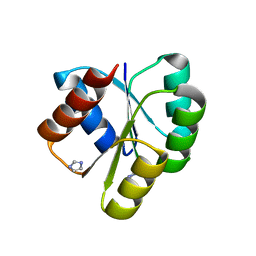 | | Crystal structure of the receiver domain of NtrX from Brucella abortus | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, IMIDAZOLE | | Authors: | Klinke, S, Fernandez, I, Carrica, M.C, Otero, L.H, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
4D3G
 
 | | Structure of PstA | | Descriptor: | PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
4CX8
 
 | | Monomeric pseudorabies virus protease pUL26N at 2.5 A resolution | | Descriptor: | PSEUDORABIES VIRUS PROTEASE | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease.
Plos Pathog., 11, 2015
|
|
6HRG
 
 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
4FPI
 
 | | Crystal Structure of 5-chloromuconolactone isomerase from Rhodococcus opacus 1CP | | Descriptor: | 5-chloromuconolactone dehalogenase | | Authors: | Ferraroni, M, Kolomytseva, M, Briganti, F, Golovleva, L.A, Scozzafava, A. | | Deposit date: | 2012-06-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and molecular docking studies on a unique chloromuconolactone dehalogenase from Rhodococcus opacus 1CP.
J.Struct.Biol., 182, 2013
|
|
4OEE
 
 | | Crystal Structure Analysis of FGF2-Disaccharide (S3I2) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
4K4D
 
 | | X-ray crystal structure of E. coli YbdB complexed with 2,4-dihydroxyphenacyl-CoA | | Descriptor: | 2,4-dihydroxyphenacyl coenzyme A, ACETATE ION, MALONATE ION, ... | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
3RH0
 
 | |
4KAX
 
 | | Crystal structure of the Grp1 PH domain in complex with Arf6-GTP | | Descriptor: | ADP-ribosylation factor 6, CITRIC ACID, Cytohesin-3, ... | | Authors: | Lambright, D.G, Malaby, A.W, van den Berg, B. | | Deposit date: | 2013-04-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F9A
 
 | | Human CDC7 kinase in complex with DBF4 and nucleotide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle 7-related protein kinase, MAGNESIUM ION, ... | | Authors: | Hughes, S, Cherepanov, P. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of human CDC7 kinase in complex with its activator DBF4.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RJS
 
 | |
3RNM
 
 | | The crystal structure of the subunit binding of human dihydrolipoamide transacylase (E2b) bound to human dihydrolipoamide dehydrogenase (E3) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-MERCAPTOETHANOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Brautigam, C.A, Wynn, R.M, Chuang, J.C, Young, B.B, Chuang, D.T. | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Basis for Weak Interactions between Dihydrolipoamide Dehydrogenase and Subunit-binding Domain of the Branched-chain {alpha}-Ketoacid Dehydrogenase Complex.
J.Biol.Chem., 286, 2011
|
|
1QFX
 
 | | PH 2.5 ACID PHOSPHATASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PROTEIN (PH 2.5 ACID PHOSPHATASE), ... | | Authors: | Kostrewa, D, Wyss, M, D'Arcy, A, Van Loon, A.P.G.M. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2. 4 A resolution.
J.Mol.Biol., 288, 1999
|
|
4KRX
 
 | | Structure of Aes from E. coli | | Descriptor: | Acetyl esterase, TETRAETHYLENE GLYCOL | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
