6P5R
 
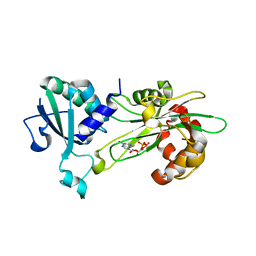 | | Structure of T. brucei MERS1-GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial edited mRNA stability factor 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-05-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
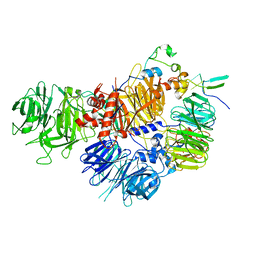
7ZGP
 
 | |
3J0K
 
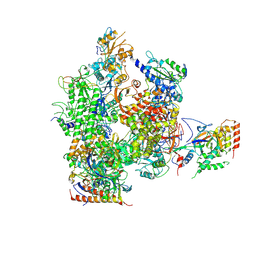 | | Orientation of RNA polymerase II within the human VP16-Mediator-pol II-TFIIF assembly | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Bernecky, C, Grob, P, Ebmeier, C.C, Nogales, E, Taatjes, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (36 Å) | | Cite: | Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly.
Plos Biol., 9, 2011
|
|
6NL1
 
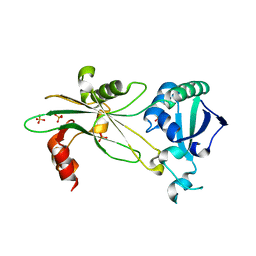 | | Structure of T. brucei MERS1 protein in its apo form | | Descriptor: | Mitochondrial edited mRNA stability factor 1, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
1KOG
 
 | | Crystal structure of E. coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase, Threonyl-tRNA synthetase mRNA, ... | | Authors: | Torres-Larrios, A, Dock-Bregeon, A.C, Romby, P, Rees, B, Sankaranarayanan, R, Caillet, J, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat.Struct.Biol., 9, 2002
|
|
2HW8
 
 | | Structure of ribosomal protein L1-mRNA complex at 2.1 resolution. | | Descriptor: | 1,4-BUTANEDIOL, 36-MER, 50S ribosomal protein L1, ... | | Authors: | Tishchenko, S, Nikonova, E, Nikulin, A, Nevskaya, N, Volchkov, S, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 2006-08-01 | | Release date: | 2006-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ribosomal protein L1-mRNA complex at 2.1 A resolution: common features of crystal packing of L1-RNA complexes.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
6KL8
 
 | | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site
To Be Published
|
|
6IVV
 
 | | Structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii with multiple surface binding regions at 1.26A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of peptide t-RNA hydrolase from Acinetobacter baumannii with multiple surface binding sites at 1.26 Angstrom resolution.
To Be Published
|
|
7CHD
 
 | | AtaT complexed with acetyl-methionyl-tRNAfMet | | Descriptor: | N-acetyltransferase domain-containing protein, RNA (77-MER) | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.804 Å) | | Cite: | Mechanism of aminoacyl-tRNA acetylation by an aminoacyl-tRNA acetyltransferase AtaT from enterohemorrhagic E. coli.
Nat Commun, 11, 2020
|
|
7YFX
 
 | | Cryo-EM structure of Hili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
3LQV
 
 | | Branch Recognition by SF3b14 | | Descriptor: | ADENINE, Pre-mRNA branch site protein p14, Splicing factor 3B subunit 1 | | Authors: | Schellenberg, M.J, MacMillan, A.M. | | Deposit date: | 2010-02-10 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Branch Recognition by SF3b14
Rna, 17, 2011
|
|
7MRL
 
 | |
7E8O
 
 | |
7L3O
 
 | |
3QSY
 
 | | Recognition of the methionylated initiator tRNA by the translation initiation factor 2 in Archaea | | Descriptor: | METHIONINE, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Translation initiation factor 2 subunit alpha, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Zelinskaya, N.V, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2011-02-22 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the archaeal translation initiation factor 2 in complex with a GTP analogue and Met-tRNAf(Met.)
J.Mol.Biol., 425, 2013
|
|
5FT9
 
 | | Arabidopsis thaliana nuclear protein-only RNase P 2 (PRORP2) | | Descriptor: | PROTEINACEOUS RNASE P 2, ZINC ION | | Authors: | Fernandez-Millan, P, Pinker, F, Schelcher, C, Gobert, A, Giege, P, Sauter, C. | | Deposit date: | 2016-01-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of Arabidopsis Nuclear Rnase P Alone and with tRNA Reveal Plasticities
To be Published
|
|
6T7K
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Wang, J, Tye, M, Payne, N.C, Mazitschek, R, Thompson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline
To Be Published
|
|
6QWS
 
 | |
4WB3
 
 | | Crystal structure of the mirror-image L-RNA/L-DNA aptamer NOX-D20 in complex with mouse C5a-desArg complement anaphylatoxin | | Descriptor: | ACETATE ION, CALCIUM ION, Complement C5, ... | | Authors: | Yatime, L, Maasch, C, Hoehlig, K, Klussmann, S, Vater, A, Andersen, G.R. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer.
Nat Commun, 6, 2015
|
|
4WB2
 
 | | Crystal structure of the mirror-image L-RNA/L-DNA aptamer NOX-D20 in complex with mouse C5a complement anaphylatoxin | | Descriptor: | ACETATE ION, CALCIUM ION, Complement C5, ... | | Authors: | Yatime, L, Maasch, C, Hoehlig, K, Klussmann, S, Vater, A, Andersen, G.R. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer.
Nat Commun, 6, 2015
|
|
4B3S
 
 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3M
 
 | | Crystal structure of the 30S ribosome in complex with compound 1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3T
 
 | | Crystal structure of the 30S ribosome in complex with compound 39 | | Descriptor: | (2S,3S,4R,5R,6R)-2-(aminomethyl)-5-azanyl-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-bis(azanyl)-2-[(2S,3R,4R,5S,6R)-3-azanyl-5-[(4-chlorophenyl)methoxy]-6-(hydroxymethyl)-4-oxidanyl-oxan-2-yl]oxy-6-oxidanyl-cyclohexyl]oxy-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-oxane-3,4-diol, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
6OTJ
 
 | |
