6W3V
 
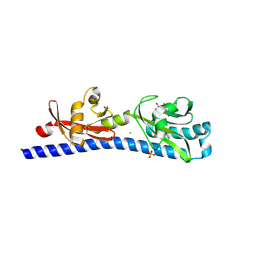 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-phenylalanine | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, PHENYLALANINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3P
 
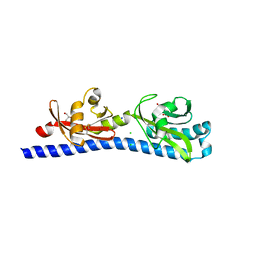 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with beta-methylnorleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3X
 
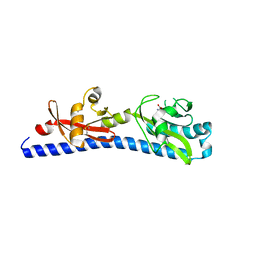 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-valine | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, SULFATE ION, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3R
 
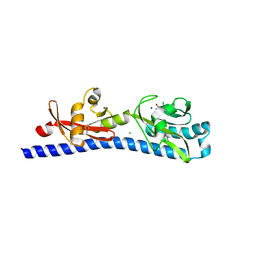 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 3-methylisoleucine | | Descriptor: | 3-methyl-L-alloisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3T
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-norvaline | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, NORVALINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
3FVG
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4J76
 
 | | Crystal Structure of a parasite tRNA synthetase, ligand-free | | Descriptor: | GLYCEROL, Tryptophanyl-tRNA synthetase | | Authors: | Koh, C.Y, Kim, J.E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structures of Plasmodium falciparum cytosolic tryptophanyl-tRNA synthetase and its potential as a target for structure-guided drug design.
Mol.Biochem.Parasitol., 189, 2013
|
|
6W3S
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-leucine | | Descriptor: | GLYCEROL, LEUCINE, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3O
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 4-methylisoleucine | | Descriptor: | 4-methylisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
1MAU
 
 | | Crystal structure of Tryptophanyl-tRNA Synthetase Complexed with ATP and Tryptophanamide in a Pre-Transition state Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, GLYCEROL, ... | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MB2
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with Tryptophan in an Open Conformation | | Descriptor: | TRYPTOPHAN, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
4E8P
 
 | |
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8R79
 
 | | The D2 domain of human DTX3L | | Descriptor: | E3 ubiquitin-protein ligase DTX3L, SULFATE ION | | Authors: | Vela-Rodriguez, C, Lehtio, L, Maksimainen, M, Duman, R, Wagner, A, Glumoff, T. | | Deposit date: | 2023-11-24 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oligomerisation mediated by the D2 domain of DTX3L is critical for DTX3L-PARP9 reading function of mono-ADP-ribosylated androgen receptor.
Biorxiv, 2023
|
|
7RC1
 
 | | X-ray Structure of SARS-CoV main protease covalently modified by compound GRL-0686 | | Descriptor: | 3C-like proteinase, 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate, DIMETHYL SULFOXIDE | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RBZ
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-017-20 | | Descriptor: | 3C-like proteinase, 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
4E8M
 
 | |
7RSM
 
 | |
5IJR
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-1 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, L-HOMOARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-29 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
1AOZ
 
 | | REFINED CRYSTAL STRUCTURE OF ASCORBATE OXIDASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Ladenstein, R, Huber, R. | | Deposit date: | 1992-01-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined crystal structure of ascorbate oxidase at 1.9 A resolution.
J.Mol.Biol., 224, 1992
|
|
7QDT
 
 | | Crystal structure of a mutant (P393GX) Thyroid Receptor Alpha ligand binding domain designed to model dominant negative human mutations. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Isoform Alpha-1 of Thyroid hormone receptor alpha | | Authors: | Romartinez-Alonso, B, Fairall, L, Agostini, M, Chatterjee, K, Schwabe, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Approach to Relieving Transcriptional Repression in Resistance to Thyroid Hormone alpha.
Mol.Cell.Biol., 42, 2022
|
|
8U1V
 
 | |
1ZVO
 
 | | Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering | | Descriptor: | Immunoglobulin delta heavy chain, myeloma immunoglobulin D lambda | | Authors: | Sun, Z, Almogren, A, Furtado, P.B, Chowdhury, B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2005-06-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-extended Solution Structure of Human Myeloma Immunoglobulin D Determined by Constrained X-ray Scattering.
J.Mol.Biol., 353, 2005
|
|
5J1X
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-5 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, Neuropilin-1, N~2~-(tert-butoxycarbonyl)-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5JHK
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-6 ligand. | | Descriptor: | N-(benzenecarbonyl)glycyl-L-arginine, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
