7D9Y
 
 | |
7DS4
 
 | |
7D8H
 
 | |
7DS3
 
 | |
7DS6
 
 | |
1W9A
 
 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
7DSB
 
 | |
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ5
 
 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
6R87
 
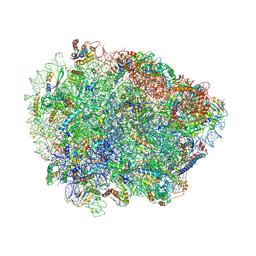 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state without Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
7D3A
 
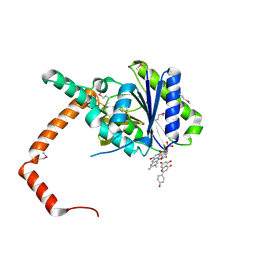 | | flavone reductase | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
6RAK
 
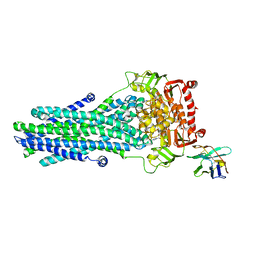 | | Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
4P6H
 
 | |
7DHP
 
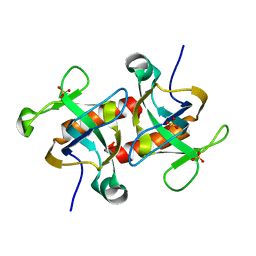 | | Crystal structure of MazF from Deinococcus radiodurans | | Descriptor: | Endoribonuclease MazF, SULFATE ION | | Authors: | Zhao, Y, Dai, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | MazEF Toxin-Antitoxin System-Mediated DNA Damage Stress Response in Deinococcus radiodurans.
Front Genet, 12, 2021
|
|
4GMT
 
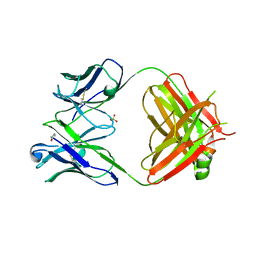 | | Crystal structure of heterosubtypic Fab S139/1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S139/1 heavy chain, ... | | Authors: | Lee, P.S, Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LRC
 
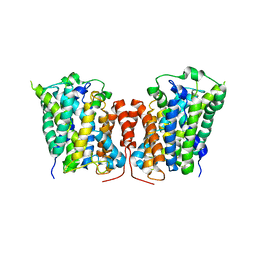 | | Structure of E. coli AdiC (P1) | | Descriptor: | Arginine/agmatine antiporter | | Authors: | Gao, X, Lu, F, Zhou, L, Shi, Y. | | Deposit date: | 2010-02-11 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structure and mechanism of an amino acid antiporter
Science, 324, 2009
|
|
7NBX
 
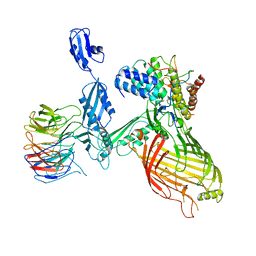 | |
6R9Z
 
 | |
7DAF
 
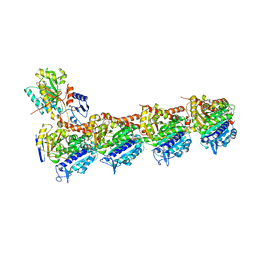 | | IXA in complex with tubulin | | Descriptor: | (1~{S},3~{S},7~{S},10~{R},11~{S},12~{S},16~{R})-8,8,10,12,16-pentamethyl-3-[(~{E})-1-(2-methyl-1,3-thiazol-4-yl)prop-1-en-2-yl]-7,11-bis(oxidanyl)-17-oxa-4-azabicyclo[14.1.0]heptadecane-5,9-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wu, C, Wang, Y. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7NP7
 
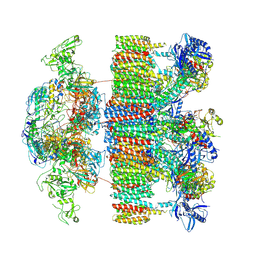 | | Structure of an intact ESX-5 inner membrane complex, Composite C1 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
7DAD
 
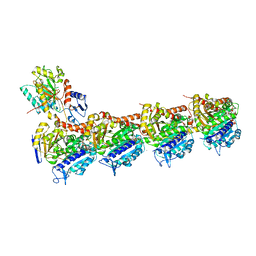 | | EPD in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y, Wu, C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7DCD
 
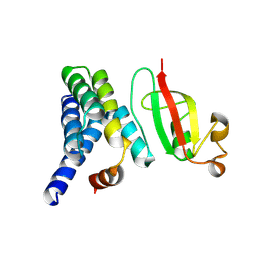 | |
7DAE
 
 | | EPB in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, CALCIUM ION, ... | | Authors: | Wu, C, Wang, Y. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6RAI
 
 | | Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding and permease protein, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
4GG8
 
 | | Immune Receptor | | Descriptor: | T-CELL RECEPTOR, SP3.4 ALPHA CHAIN, SP3.4 BETA CHAIN, ... | | Authors: | Broughton, S.E, Theodossis, A, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease.
Immunity, 37, 2012
|
|
