2R0V
 
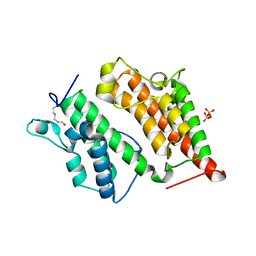 | | Structure of the Rsc4 tandem bromodomain acetylated at K25 | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, SULFATE ION | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
3QEU
 
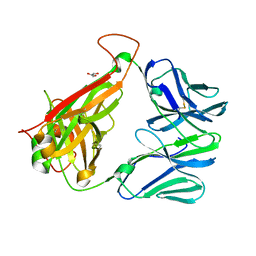 | | The crystal structure of TCR DMF5 | | Descriptor: | DMF5 alpha chain, DMF5 beta chain, GLYCEROL, ... | | Authors: | Borbulevych, O.Y, Santhanagopolan, S.M, Baker, B.M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-07-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | TCRs Used in Cancer Gene Therapy Cross-React with MART-1/Melan-A Tumor Antigens via Distinct Mechanisms.
J.Immunol., 187, 2011
|
|
3HHP
 
 | |
4KY8
 
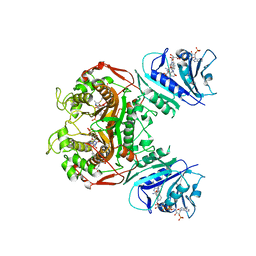 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, methotrexate, FdUMP and 4-((2-amino-6-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)thio)-2-chlorophenyl)-L-glutamic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional thymidylate synthase-dihydrofolate reductase, METHOTREXATE, ... | | Authors: | Kumar, V.P, Anderson, K.S. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Substituted pyrrolo[2,3-d]pyrimidines as Cryptosporidium hominis thymidylate synthase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2QDP
 
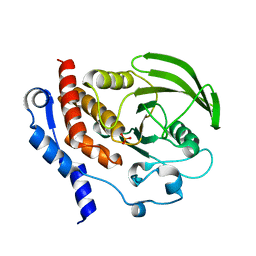 | |
4KXV
 
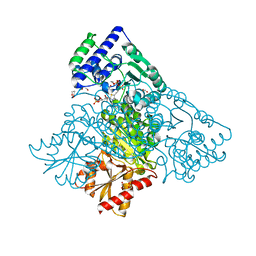 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4P98
 
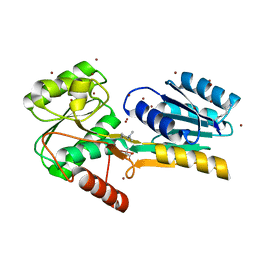 | |
2FD4
 
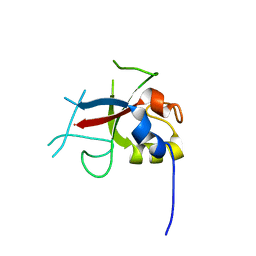 | | Crystal Structure of AvrPtoB (436-553) | | Descriptor: | avirulence protein AvrptoB | | Authors: | Janjusevic, R, Stebbins, C.E. | | Deposit date: | 2005-12-13 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase.
Science, 311, 2006
|
|
4KZ5
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 5 (N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine) | | Descriptor: | Beta-lactamase, N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
3HF7
 
 | | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A | | Descriptor: | ADENOSINE MONOPHOSPHATE, uncharacterized CBS-domain protein | | Authors: | Stein, A.J, Nocek, B, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A
To be Published
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
2R4J
 
 | | Crystal structure of Escherichia coli SeMet substituted Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3HFG
 
 | |
4L45
 
 | | Crystal structures of human p70S6K1-T389E | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
2QIO
 
 | |
3HKK
 
 | |
3HIS
 
 | |
3HJ5
 
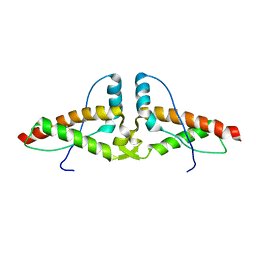 | | Human prion protein variant V129 domain swapped dimer | | Descriptor: | Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-20 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HII
 
 | |
2QIH
 
 | |
3HQY
 
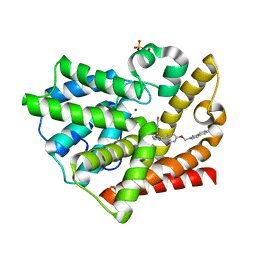 | | Discovery of novel inhibitors of PDE10A | | Descriptor: | 2-({4-[4-(pyridin-4-ylmethyl)-1H-pyrazol-3-yl]phenoxy}methyl)quinoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2009-06-08 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Novel Class of Phosphodiesterase 10A Inhibitors and Identification of Clinical Candidate 2-[4-(1-Methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the Treatment of Schizophrenia
J.Med.Chem., 52, 2009
|
|
3HR8
 
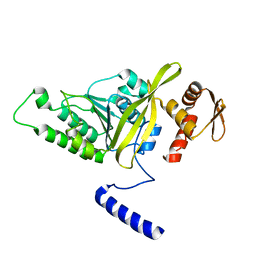 | | Crystal Structure of Thermotoga maritima RecA | | Descriptor: | Protein recA | | Authors: | Lee, S, Kim, T.G, Jeong, E.-Y, Ban, C, Jeon, W.-J, Min, K.I, Song, K.-M, Heo, S.-D, Ku, J.K. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of RecA Protein from Thermotoga maritima MSB8
to be published
|
|
2QON
 
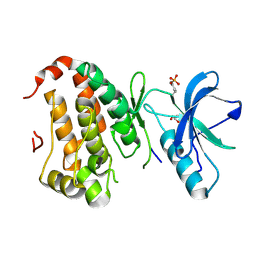 | | Human EphA3 kinase and juxtamembrane region, Y596F:Y602F:Y742A triple mutant | | Descriptor: | Ephrin receptor, GLYCEROL | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QK0
 
 | |
4LB3
 
 | | Crystal structure of human AR complexed with NADP+ and {5-chloro-2-[(2-fluoro-4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-chloro-2-[(2-fluoro-4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
