2HUO
 
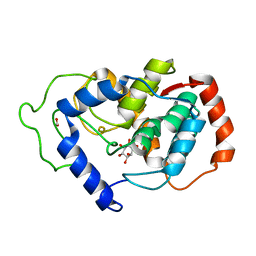 | | Crystal structure of mouse myo-inositol oxygenase in complex with substrate | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Brown, P.M, Caradoc-Davies, T.T, Dickson, J.M.J, Cooper, G.J.S, Loomes, K.M, Baker, E.N. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DWV
 
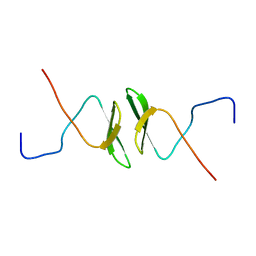 | | Solution structure of the second WW domain from mouse salvador homolog 1 protein (mWW45) | | Descriptor: | Salvador homolog 1 protein | | Authors: | Ohnishi, S, Kigawa, T, Koshiba, S, Tomizawa, T, Sato, M, Tochio, N, Inoue, M, Harada, T, Watanabe, S, Guntert, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-17 | | Release date: | 2007-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form
Febs Lett., 581, 2007
|
|
4FHE
 
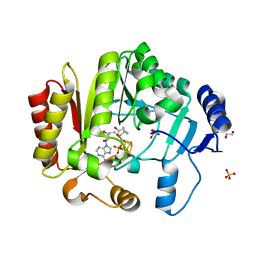 | | Spore photoproduct lyase C140A mutant | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
2QJI
 
 | | M. jannaschii ADH synthase complexed with dihydroxyacetone phosphate and glycerol | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
2ZWN
 
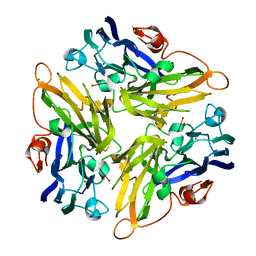 | | Crystal structure of the novel two-domain type laccase from a metagenome | | Descriptor: | CHLORIDE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2008-12-17 | | Release date: | 2009-04-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of a two-domain type laccase: a missing link in the evolution of multi-copper proteins
Febs Lett., 583, 2009
|
|
2QMC
 
 | |
3FFK
 
 | | Crystal structure of human Gelsolin domains G1-G3 bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, actin, ... | | Authors: | Chumnarnsilpa, S, Robinson, R.C, Burtnick, L.D. | | Deposit date: | 2008-12-03 | | Release date: | 2009-10-06 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2DV6
 
 | | Crystal structure of nitrite reductase from Hyphomicrobium denitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, POTASSIUM ION | | Authors: | Nojiri, M, Xie, Y, Yamamoto, T, Inoue, T, Suzuki, S, Kai, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a hexameric copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4CHL
 
 | | Human Ethylmalonic Encephalopathy Protein 1 (hETHE1) | | Descriptor: | FE (III) ION, PERSULFIDE DIOXYGENASE ETHE1, MITOCHONDRIAL | | Authors: | Pettinati, I, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-12-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human persulfide dioxygenase: structural basis of ethylmalonic encephalopathy.
Hum. Mol. Genet., 24, 2015
|
|
2YGW
 
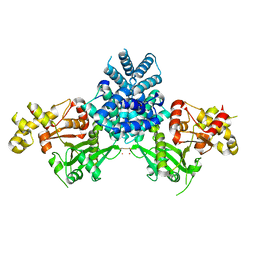 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
2JBF
 
 | |
2ZR8
 
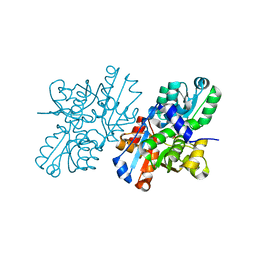 | | Crystal Structure of Modified Serine Racemase complexed with Serine | | Descriptor: | MAGNESIUM ION, N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, SERINE, ... | | Authors: | Goto, M. | | Deposit date: | 2008-08-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe
J.Biol.Chem., 284, 2009
|
|
2ERR
 
 | |
2ERO
 
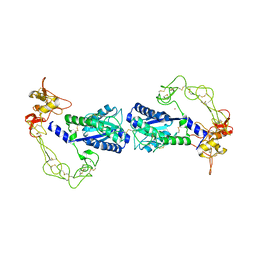 | | Crystal structure of vascular apoptosis-inducing protein-1(orthorhombic crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COBALT (III) ION, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2005-10-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold
Embo J., 25, 2006
|
|
3LIS
 
 | |
7ZA4
 
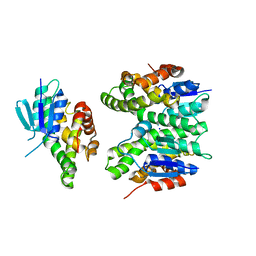 | | GSTF sh155 mutant | | Descriptor: | Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
3CVL
 
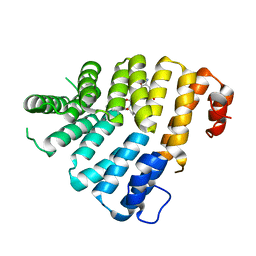 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVQ
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
7ZA5
 
 | | GSTF sh101 mutant | | Descriptor: | Glutathione transferase | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
3D2U
 
 | | Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Actin, Beta-2-microglobulin, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2008-05-08 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3H1T
 
 | |
2ERP
 
 | | Crystal structure of vascular apoptosis-inducing protein-1(inhibitor-bound form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, CALCIUM ION, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2005-10-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold
Embo J., 25, 2006
|
|
4NMI
 
 | |
6A5I
 
 | | Pseudocerastes Persicus Trypsin Inhibitor | | Descriptor: | Trypsin Inhibitor | | Authors: | Amininasab, M. | | Deposit date: | 2018-06-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of PPTI, a kunitz-type protein from the venom of Pseudocerastes persicus.
PLoS ONE, 14, 2019
|
|
3SF0
 
 | |
