3HJ6
 
 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
5RPW
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo63 | | Descriptor: | Proteinase K | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3HES
 
 | | Human prion protein variant F198S with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
2HLQ
 
 | |
3HFE
 
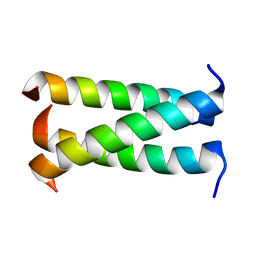 | | A trimeric form of the Kv7.1 A domain Tail | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Xu, Q, Minor, D.L. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a trimeric form of the K(V)7.1 (KCNQ1) A-domain tail coiled-coil reveals structural plasticity and context dependent changes in a putative coiled-coil trimerization motif.
Protein Sci., 18, 2009
|
|
3HG2
 
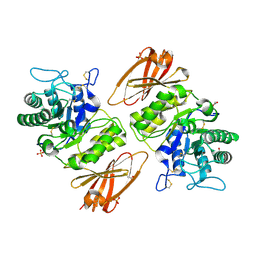 | | Human alpha-galactosidase catalytic mechanism 1. Empty active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Alpha-galactosidase A, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
4KXX
 
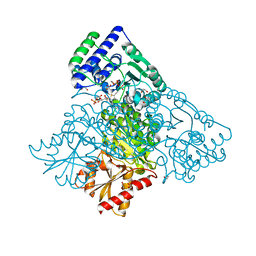 | | Human transketolase in covalent complex with donor ketose D-sedoheptulose-7-phosphate | | Descriptor: | (2R,3R,4S,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl dihydrogen phosphate, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
2OUK
 
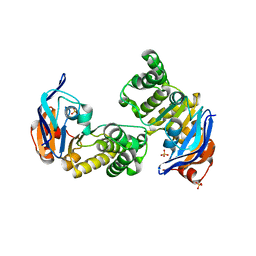 | | ABC Protein ArtP in complex with Sulphate | | Descriptor: | SULFATE ION, protein ArtP | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2007-02-11 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the ATP-binding cassette (ABC) protein ArtP from Geobacillus stearothermophilus reveal a stable dimer in the post hydrolysis state and an asymmetry in the dimerization region
To be Published
|
|
4KZ6
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 13 ((2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid) | | Descriptor: | (2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZA
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 48 (3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid) | | Descriptor: | 3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4L04
 
 | |
4L0G
 
 | | Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside hydrolase family 48, ... | | Authors: | Jiao, A, Bai, A, Yan, F, Geng, W. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary X-ray analysis of a processive cellobiohydrolase CbCBH48A from Caldicellulosiruptor bescii
to be published
|
|
3HIT
 
 | | Crystal structure of Saporin-L1 in complex with the dinucleotide inhibitor, a transition state analogue | | Descriptor: | 5'-O-[(S)-{[(3R,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[(S)-hydroxy(3-hydroxypropoxy)phosphoryl]oxy}methyl)pyrrolidin-3-yl]oxy}(hydroxy)phosphoryl]-3'-O-[(R)-hydroxy(4-hydroxybutoxy)phosphoryl]-2'-O-methylguanosine, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2OWY
 
 | |
2E4G
 
 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
3HE5
 
 | |
2OYA
 
 | | Crystal structure analysis of the dimeric form of the SRCR domain of mouse MARCO | | Descriptor: | Macrophage receptor MARCO, SULFATE ION | | Authors: | Ojala, J.R.M, Pikkarainen, T, Tuuttila, A, Sandalova, T, Tryggvason, K. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition.
J.Biol.Chem., 282, 2007
|
|
2HLJ
 
 | |
2OZP
 
 | |
2OLX
 
 | |
3HGF
 
 | | Expression, purification, spectroscopical and crystallographical studies of segments of the nucleotide binding domain of the reticulocyte binding protein Py235 of Plasmodium yoelii | | Descriptor: | Rhoptry protein fragment | | Authors: | Gruber, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Jeyakanthan, J, Preiser, P.R, Gruber, G. | | Deposit date: | 2009-05-13 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural determination of functional units of the nucleotide binding domain (NBD94) of the reticulocyte binding protein Py235 of Plasmodium yoelii
Plos One, 5, 2010
|
|
2OMX
 
 | | Crystal structure of InlA S192N G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ONA
 
 | |
3HHC
 
 | |
