4V24
 
 | | Sphingosine kinase 1 in complex with PF-543 | | Descriptor: | ACETATE ION, SPHINGOSINE KINASE 1, {(2R)-1-[4-({3-METHYL-5-[(PHENYLSULFONYL)METHYL]PHENOXY}METHYL)BENZYL]PYRROLIDIN-2-YL}METHANOL | | Authors: | Elkins, J.M, Wang, J, Sorrell, F, Tallant, C, Wang, D, Shrestha, L, Bountra, C, von Delft, F, Knapp, S, Edwards, A. | | Deposit date: | 2014-10-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sphingosine Kinase 1 with Pf-543.
Acs Med.Chem.Lett., 5, 2014
|
|
9MEA
 
 | |
4WAF
 
 | | Crystal Structure of a novel tetrahydropyrazolo[1,5-a]pyrazine in an engineered PI3K alpha | | Descriptor: | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Drug Design of Novel Potent and Selective Tetrahydropyrazolo[1,5-a]pyrazines as ATR Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
6J7A
 
 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (17aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.269 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
6J7I
 
 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (15aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
9JZB
 
 | | PfDXR - Mn2+ - MAMK251 ternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
9JZE
 
 | | PfDXR - Mn2+ - NADPH - MAMK431 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
9JZD
 
 | | PfDXR - Mn2+ - NADPH - MAMK433 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
6JMS
 
 | | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen | | Descriptor: | 1,2-ETHANEDIOL, Pollen allergen CJP38 | | Authors: | Takashima, T, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen
To Be Published
|
|
9JZA
 
 | | PfDXR - Mn2+ - NADPH - MAMK218 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
9JZC
 
 | | PfDXR - Mn2+ - NADPH - MAMK251 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
9VK1
 
 | | Structure of the plant diacylglycerol O-acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | Diacylglycerol O-acyltransferase 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Liu, X.Y, Li, J.J, Song, D.F, Liu, Z.F. | | Deposit date: | 2025-06-22 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural mechanisms underlying the free fatty acid-mediated regulation of DIACYLGLYCEROL O-ACYLTRANSFERASE 1 in Arabidopsis.
Plant Cell, 37, 2025
|
|
4W88
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with a xyloglucan oligosaccharide and TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, ... | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4V28
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with Man-Man-Methylumbelliferone | | Descriptor: | 1,2-ETHANEDIOL, 7-hydroxy-4-methyl-2H-chromen-2-one, ACETATE ION, ... | | Authors: | Hakki, Z, Bellmaine, S, Thompson, A.J, Speciale, G, Davies, G.J, Williams, S.J. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Dissection of the Endo-Alpha-1,2-Mannanase Activity of Bacterial Gh99 Glycoside Hydrolases from Bacteroides Spp.
Chemistry, 21, 2015
|
|
4UYD
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 1,3-dimethyl-2-oxo-2,3- dihydro-1H-1,3-benzodiazole-5-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxamide, BROMODOMAIN-CONTAINING PROTEIN 4, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2014-08-30 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | 1,3-Dimethyl Benzimidazolones are Potent, Selective Inhibitors of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 5, 2014
|
|
4UYE
 
 | | BROMODOMAIN OF HUMAN BRPF1 WITH N-1,3-dimethyl-2-oxo-6-(piperidin-1- yl)-2,3-dihydro-1H-1,3-benzodiazol-5-yl-2-methoxybenzamide | | Descriptor: | 1,2-ETHANEDIOL, N-[1,3-dimethyl-2-oxo-6-(piperidin-1-yl)-2,3-dihydro-1H-benzimidazol-5-yl]-2-methoxybenzamide, PEREGRIN | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2014-08-30 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1,3-Dimethyl Benzimidazolones are Potent, Selective Inhibitors of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 5, 2014
|
|
4W86
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose and TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, ... | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4WH2
 
 | | N-acetylhexosamine 1-kinase in complex with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WKI
 
 | |
4WKE
 
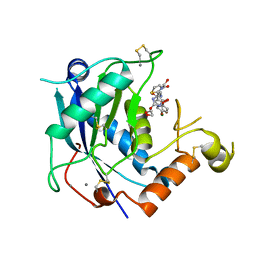 | | Crystal structure of human ADAMTS-4 in complex with inhibitor 5-chloro-N-{[(4R)-2,5-dioxo-4-(1,3-thiazol-2-yl)imidazolidin-4-yl]methyl}-1-benzofuran-2-carboxamide (compound 10) | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-{[(4R)-2,5-dioxo-4-(1,3-thiazol-2-yl)imidazolidin-4-yl]methyl}-1-benzofuran-2-carboxamide, A disintegrin and metalloproteinase with thrombospondin motifs 4, ... | | Authors: | Durbin, J.D. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of potent and selective hydantoin inhibitors of aggrecanase-1 and aggrecanase-2 that are efficacious in both chemical and surgical models of osteoarthritis.
J.Med.Chem., 57, 2014
|
|
4WWP
 
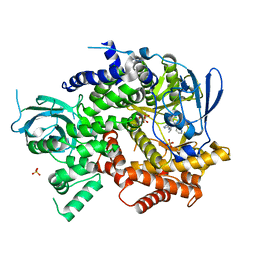 | | Crystal structure of human PI3K-gamma in complex with pyridinylquinoline inhibitor N-{(1S)-1-[8-chloro-2-(2-methylpyridin-3-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine | | Descriptor: | GLYCEROL, N-{(1S)-1-[8-chloro-2-(2-methylpyridin-3-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, ... | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
4WH3
 
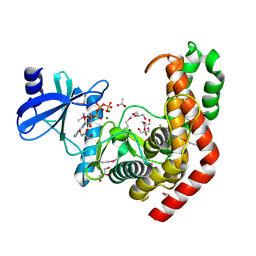 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WSY
 
 | |
4WT6
 
 | |
