1KN4
 
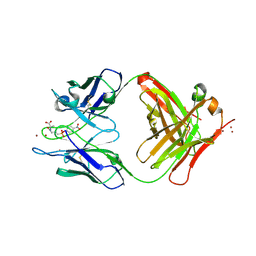 | | CATALYTIC ANTIBODY D2.3 COMPLEX | | Descriptor: | IG ANTIBODY D2.3 (HEAVY CHAIN), IG ANTIBODY D2.3 (LIGHT CHAIN), PARA-NITROPHENYL PHOSPHONOBUTANOYL D-ALANINE, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remarkable remote chiral recognition in a reaction mediated by a catalytic antibody.
J.Am.Chem.Soc., 124, 2002
|
|
1KN5
 
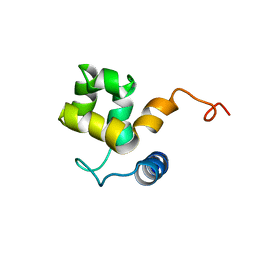 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1KN6
 
 | |
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
1KN9
 
 | |
1KNA
 
 | |
1KNB
 
 | |
1KNC
 
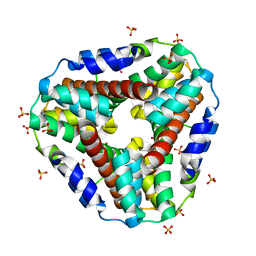 | | Structure of AhpD from Mycobacterium tuberculosis, a novel enzyme with thioredoxin-like activity. | | Descriptor: | AhpD protein, SULFATE ION | | Authors: | Bryk, R, Lima, C.D, Erdjument-Bromage, H, Tempst, P, Nathan, C. | | Deposit date: | 2001-12-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein.
Science, 295, 2002
|
|
1KND
 
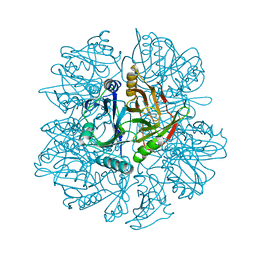 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, CATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
1KNE
 
 | |
1KNF
 
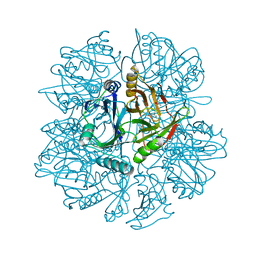 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 3-methyl Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, 3-METHYLCATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
1KNG
 
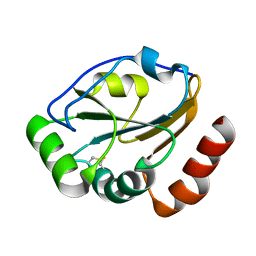 | | Crystal structure of CcmG reducing oxidoreductase at 1.14 A | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN CYCY | | Authors: | Edeling, M.A, Guddat, L.W, Fabianek, R.A, Thony-Meyer, L, Martin, J.L. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment
Structure, 10, 2002
|
|
1KNI
 
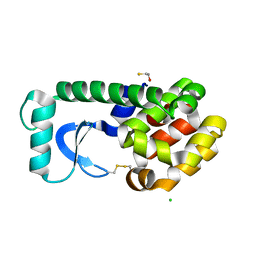 | | Stabilizing Disulfide Bridge Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Jacobson, R.H, Matsumura, M, Faber, H.R, Matthews, B.W. | | Deposit date: | 2001-12-18 | | Release date: | 2001-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a stabilizing disulfide bridge mutant that closes the active-site cleft of T4 lysozyme.
Protein Sci., 1, 1992
|
|
1KNJ
 
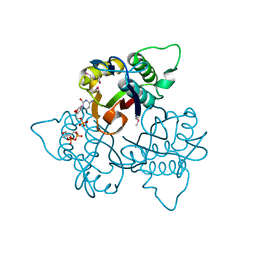 | | Co-Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli Involved in Mevalonate-Independent Isoprenoid Biosynthesis, Complexed with CMP/MECDP/Mn2+ | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1KNK
 
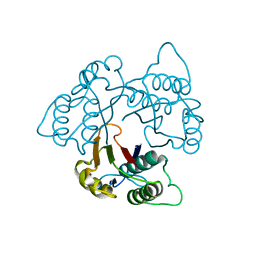 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1KNL
 
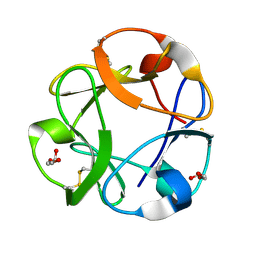 | | Streptomyces lividans Xylan Binding Domain cbm13 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KNM
 
 | | Streptomyces lividans Xylan Binding Domain cbm13 in Complex with Lactose | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KNO
 
 | |
1KNP
 
 | |
1KNQ
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KNR
 
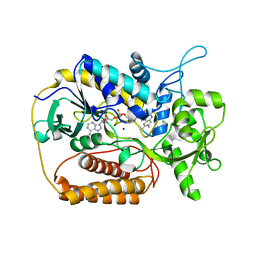 | | L-aspartate oxidase: R386L mutant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase, ... | | Authors: | Bossi, R.T, Mattevi, A. | | Deposit date: | 2001-12-19 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis.
Biochemistry, 41, 2002
|
|
1KNT
 
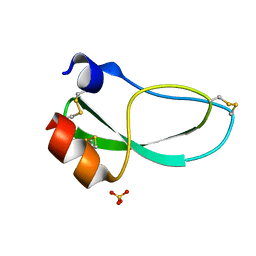 | | THE 1.6 ANGSTROMS STRUCTURE OF THE KUNITZ-TYPE DOMAIN FROM THE ALPHA3 CHAIN OF THE HUMAN TYPE VI COLLAGEN | | Descriptor: | COLLAGEN TYPE VI, SULFATE ION | | Authors: | Arnoux, B, Merigeau, K, Saludjian, P, Norris, F, Norris, K, Bjorn, S, Olsen, O, Petersen, L, Ducruix, A. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A structure of Kunitz-type domain from the alpha 3 chain of human type VI collagen.
J.Mol.Biol., 246, 1995
|
|
1KNU
 
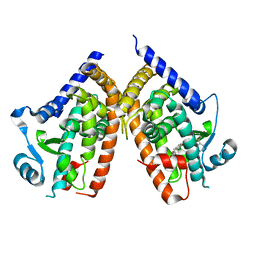 | | LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA IN COMPLEX WITH A SYNTHETIC AGONIST | | Descriptor: | (S)-3-(4-(2-CARBAZOL-9-YL-ETHOXY)-PHENYL)-2-ETHOXY-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Svensson, L.A, Mortensen, S.B, Fleckner, J, Woeldike, H.F. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclic-alpha-alkyloxyphenylpropionic acids: dual PPARalpha/gamma agonists with hypolipidemic and antidiabetic activity
J.MED.CHEM., 45, 2002
|
|
1KNV
 
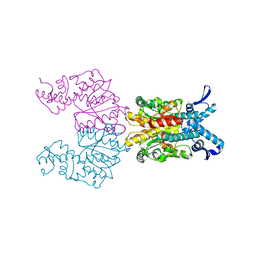 | | Bse634I restriction endonuclease | | Descriptor: | ACETATE ION, Bse634I restriction endonuclease, CHLORIDE ION | | Authors: | Grazulis, S, Deibert, M, Rimseliene, R, Skirgaila, R, Sasnauskas, G, Lagunavicius, A, Repin, V, Urbanke, C, Huber, R, Siksnys, V. | | Deposit date: | 2001-12-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence.
Nucleic Acids Res., 30, 2002
|
|
1KNW
 
 | | Crystal structure of diaminopimelate decarboxylase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Diaminopimelate decarboxylase, LITHIUM ION, ... | | Authors: | Levdikov, V, Blagova, L, Bose, N, Momany, C. | | Deposit date: | 2001-12-19 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diaminopimelate Decarboxylase uses a Versatile Active Site for Stereospecific Decarboxylation
To be Published
|
|
