4E0X
 
 | |
4DE3
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 4 | | Descriptor: | 3-bromo-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4DEC
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate (UDP) and phosphoglyceric acid (PGA) | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, ... | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
4DEW
 
 | |
4DIZ
 
 | |
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
4DJU
 
 | | Structure of BACE Bound to 2-imino-3-methyl-5,5-diphenylimidazolidin-4-one | | Descriptor: | (2E)-2-imino-3-methyl-5,5-diphenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DKV
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-10007 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2012-02-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1847 Å) | | Cite: | Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study.
Antimicrob. Agents Chemother., 58, 2014
|
|
4DLV
 
 | | H-Ras Set 2 CaCl2/DTT, ordered off | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, GTPase HRas, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
4DN1
 
 | | Crystal structure of an ENOLASE (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isomerase/lactonizing enzyme, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Sojitra, S, Al Obaidi, N.F, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate
to be published
|
|
4DJX
 
 | |
4DKU
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-09027 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2012-02-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4902 Å) | | Cite: | Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study.
Antimicrob. Agents Chemother., 58, 2014
|
|
4DPP
 
 | | The structure of dihydrodipicolinate synthase 2 from Arabidopsis thaliana | | Descriptor: | Dihydrodipicolinate synthase 2, chloroplastic, SODIUM ION | | Authors: | Griffin, M.D.W, Billakanti, J.M, Gerrard, J.A, Dobson, R.C.J, Pearce, F.G. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterisation of the first enzymes committed to lysine biosynthesis in Arabidopsis thaliana
Plos One, 7, 2012
|
|
4FDW
 
 | |
4FMR
 
 | |
4FHO
 
 | |
4F53
 
 | |
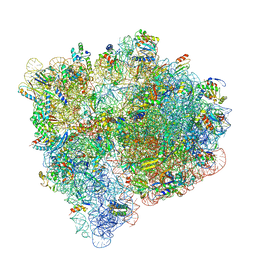
7JT3
 
 | |
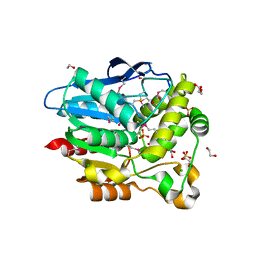
4F0J
 
 | |
4F3T
 
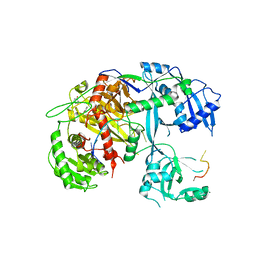 | | Human Argonaute-2 - miR-20a complex | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*UP*AP*AP*AP*GP*UP*GP*CP*UP*UP*AP*UP*AP*GP*UP*G*CP*AP*GP*G)-3') | | Authors: | Elkayam, E, Kuhn, C.-D, Tocilj, A, Joshua-Tor, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Human Argonaute-2 in Complex with miR-20a.
Cell(Cambridge,Mass.), 150, 2012
|
|
4EH9
 
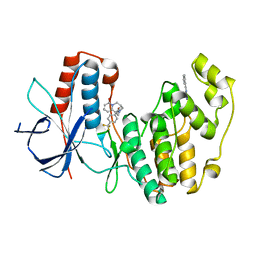 | | Human p38 MAP kinase in complex with NP-F11 and RL87 | | Descriptor: | (1R,5S)-3-[4-(trifluoromethyl)benzoyl]-1,2,3,4,5,6-hexahydro-8H-1,5-methanopyrido[1,2-a][1,5]diazocin-8-one, Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
4ESV
 
 | | A New Twist on the Translocation Mechanism of Helicases from the Structure of DnaB with its Substrates | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Itsathitphaisarn, O, Wing, R.A, Eliason, W.K, Wang, J, Steitz, T.A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation.
Cell(Cambridge,Mass.), 151, 2012
|
|
4E4G
 
 | | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
4E6M
 
 | | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium)
To be Published
|
|
4E9K
 
 | |
