4ALO
 
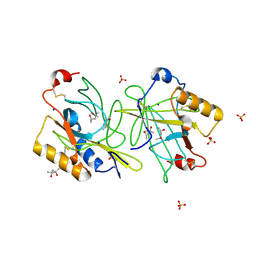 | | STRUCTURE AND PROPERTIES OF H1 CRUSTACYANIN FROM LOBSTER HOMARUS AMERICANUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, H1 APOCRUSTACYANIN, SODIUM ION, ... | | Authors: | Ferrari, M, Folli, C, Pincolini, E, Mcclintock, T.S, Roessle, M, Berni, R, Cianci, M. | | Deposit date: | 2012-03-05 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Recombinant Crustacyanin Subunits from the Lobster Homarus Americanus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8GEW
 
 | | H-FABP crystal soaked in a bromo palmitic acid solution | | Descriptor: | 2-Bromopalmitic acid, Fatty acid-binding protein, heart, ... | | Authors: | Howard, E, Cousido-Siah, A, Alvarez, A, Espinosa, Y, Podjarny, A, Mitschler, A, Carlevaro, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lipid exchange in crystal-confined fatty acid binding proteins: X-ray evidence and molecular dynamics explanation.
Proteins, 91, 2023
|
|
8GEM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethanamine | | Descriptor: | N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GDM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with {[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}(methyl)[1-(thiophen-2-yl)ethyl]amine | | Descriptor: | (1S)-N-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-N-methyl-1-(thiophen-2-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEY
 
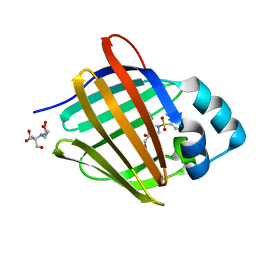 | | Crystal structure of human cellular retinol binding protein 1 in complex with 4-(hydroxymethyl)-1-[(4-methoxy-5,6,7,8-tetrahydronaphthalen-1-yl)sulfonyl]piperidin-4-ol | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(hydroxymethyl)-1-(4-methoxy-5,6,7,8-tetrahydronaphthalene-1-sulfonyl)piperidin-4-ol, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GD2
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-(2-thienylmethyl)methanamine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(thiophen-2-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEV
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine | | Descriptor: | 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEU
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with methyl({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)[(1-methylpyrazol-4-yl)methyl]amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(1-methyl-1H-pyrazol-4-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8HTA
 
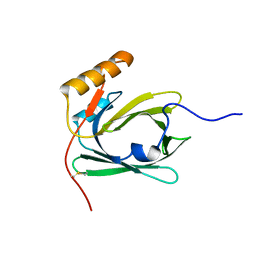 | |
8IVL
 
 | | FABP7 complexed with Cholesterol | | Descriptor: | CHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
8IVF
 
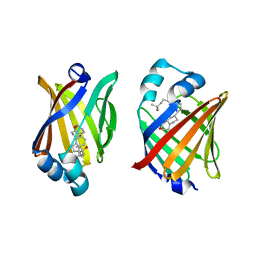 | | FABP7 complexed with 25-HC | | Descriptor: | 25-HYDROXYCHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
2CZU
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
2CZT
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
5DPQ
 
 | | Crystal Structure of E72A mutant of domain swapped dimer Human Cellular Retinol Binding Protein | | Descriptor: | ACETATE ION, Retinol-binding protein 2 | | Authors: | Assar, Z, Nossoni, Z, Wang, W, Geiger, J.H, Borhan, B. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
5EDC
 
 | |
6RYT
 
 | |
5F58
 
 | |
6RWP
 
 | |
6RWQ
 
 | | Engineered beta-lactoglobulin: variant F105L in complex with myristic acid | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, MYRISTIC ACID | | Authors: | Loch, J.I, Gotkowski, M, Lewinski, K. | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design approach to rational site-directed mutagenesis of beta-lactoglobulin.
J.Struct.Biol., 210, 2020
|
|
6RWR
 
 | |
5F6B
 
 | |
2CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-13 6307) | | Descriptor: | 3-METHYL-7-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL) -OCTA-2,4,6-TRIENOIC ACID, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6S2S
 
 | |
2DM5
 
 | | Thermodynamic Penalty Arising From Burial of a Ligand Polar Group Within a Hydrophobic Pocket of a Protein Receptor | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTANE-1,8-DIOL | | Authors: | Barratt, E, Bronowska, A, Vondrasek, J, Bingham, R, Phillips, S, Homans, S.W. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor
J.Mol.Biol., 362, 2006
|
|
5FAZ
 
 | |
