2CHR
 
 | |
1MNS
 
 | |
1MRA
 
 | |
3DG6
 
 | | Crystal structure of muconate lactonizing enzyme from Mucobacterium Smegmatis complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3DG7
 
 | | Crystal structure of muconate lactonizing enzyme from Mucobacterium Smegmatis complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
4V6F
 
 | | Elongation complex of the 70S ribosome with three tRNAs and mRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 23S RRNA, ... | | Authors: | Jenner, L.B, Yusupova, G, Yusupov, M. | | Deposit date: | 2009-07-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2HG8
 
 | |
4M6U
 
 | |
1JPD
 
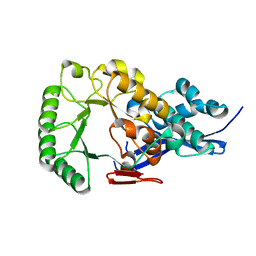 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | L-Ala-D/L-Glu Epimerase | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
1MDL
 
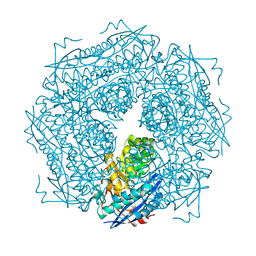 | |
1MDR
 
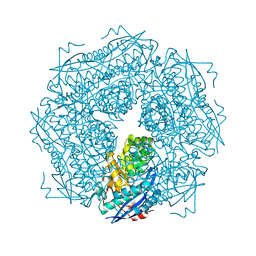 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | Authors: | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | Deposit date: | 1993-11-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
3BJS
 
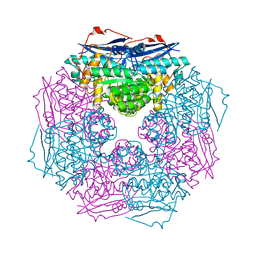 | | Crystal structure of a member of enolase superfamily from Polaromonas sp. JS666 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Dickey, M, Sauder, J.M, Reyes, C, Groshong, C, Gheyi, T, Smith, D, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Polaromonas sp. JS666.
To be Published
|
|
3I6T
 
 | | Crystal structure of muconate cycloisomerase from Jannaschia sp. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, POTASSIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of muconate cycloisomerase from Jannaschia sp.
To be Published
|
|
3I6E
 
 | | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM Ruegeria pomeroyi. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase I, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of muconate lactonizing enzyme from Ruegeria pomeroyi.
To be Published
|
|
3MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT I54V | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-27 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
3MY9
 
 | | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Quartararo, C.E, Ramagopal, U, Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans
To be Published
|
|
1DTN
 
 | |
4V9Q
 
 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | Deposit date: | 2013-06-12 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V88
 
 | | The structure of the eukaryotic ribosome at 3.0 A resolution. | | Descriptor: | 18S RIBOSOMAL RNA, 18S rRNA, 25S rRNA, ... | | Authors: | Ben-Shem, A, Garreau de Loubresse, N, Melnikov, S, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2011-10-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the eukaryotic ribosome at 3.0 angstrom resolution.
Science, 334, 2011
|
|
3VCN
 
 | | Crystal structure of mannonate dehydratase (target EFI-502209) from Caulobacter crescentus CB15 | | Descriptor: | CARBONATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of mannonate dehydratase from Caulobacter crescentus CB15
To be Published
|
|
2GSH
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium | | Descriptor: | GLYCEROL, L-RHAMNONATE DEHYDRATASE, MAGNESIUM ION | | Authors: | Patskovsky, Y, Malashkevich, V.N, Sauder, J.M, Dickey, M, Adams, J.M, Wasserman, S.R, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal Structure of L-rhamnonate dehydratase from Salmonella Typhimurium Lt2
To be Published
|
|
4E4F
 
 | | Crystal structure of enolase PC1_0802 (TARGET EFI-502240) from Pectobacterium carotovorum subsp. carotovorum PC1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ENOLASE PC1_0802 from Pectobacterium carotovorum
To be Published
|
|
4FP1
 
 | | P. putida mandelate racemase co-crystallized with 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl) propionic acid | | Descriptor: | 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propanoic acid, MAGNESIUM ION, Mandelate racemase | | Authors: | Lietzan, A.D, St.Maurice, M. | | Deposit date: | 2012-06-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Potent inhibition of mandelate racemase by a fluorinated substrate-product analogue with a novel binding mode.
Biochemistry, 53, 2014
|
|
4HNC
 
 | |
4IHC
 
 | | Crystal structure of probable mannonate dehydratase Dd703_0947 (target EFI-502222) from Dickeya dadantii Ech703 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mannonate dehydratase Dd703_0947 from Dickeya dadantii Ech703
To be Published
|
|
