5JQI
 
 | |
6Q0J
 
 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
6TRG
 
 | | Salmonella typhimurium neuraminidase mutant (D100S) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D100S)
To Be Published
|
|
6GAN
 
 | | Structure of fully reduced Hydrogenase (Hyd-2) variant E14Q | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
4RED
 
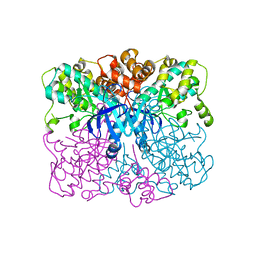 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
8E79
 
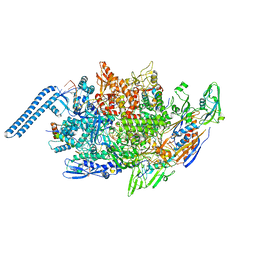 | | Mycobacterium tuberculosis RNAP paused elongation complex with Escherichia coli NusG transcription factor | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
8E95
 
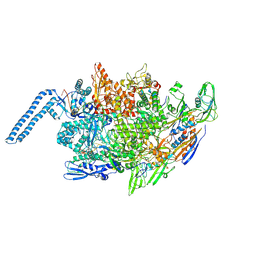 | | Mycobacterium tuberculosis RNAP elongation complex | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
8E8M
 
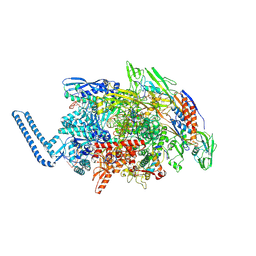 | | Mycobacterium tuberculosis RNAP paused elongation complex | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
7QUM
 
 | | ATAD2 in complex with FragLite2 | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
6NJL
 
 | |
1PTZ
 
 | | Crystal structure of the human CU, Zn Superoxide Dismutase, Familial Amyotrophic Lateral Sclerosis (FALS) Mutant H43R | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Craig, L, Huff, M.E, Thayer, M.M, Cardoso, R.M.F, Kassmann, C.J, Lo, T.P, Bruns, C.K, Powers, E.T, Kelly, J.W, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates Via Framework Destabilization
J.Mol.Biol., 332, 2003
|
|
7QZM
 
 | | ATAD2 in complex with FragLite28 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1-(2-hydroxyethyl)pyridin-2(1H)-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-31 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QYK
 
 | | ATAD2 in complex with FragLite18 | | Descriptor: | (4-bromanylpyridin-2-yl)methanol, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5KAH
 
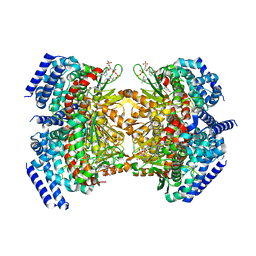 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, V425T mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.779 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7UTI
 
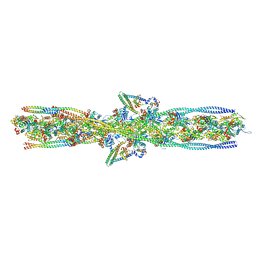 | | ALTERNATIVE MODELING OF TROPOMYOSIN IN HUMAN CARDIAC THIN FILAMENT IN THE CALCIUM BOUND STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rynkiewicz, M.J, Pavadai, E, Lehman, W. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Biophys J, 119, 2020
|
|
5WEW
 
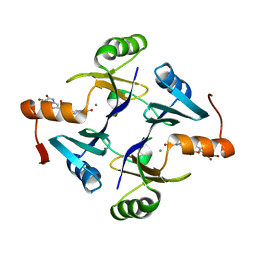 | | Crystal structure of Klebsiella pneumoniae fosfomycin resistance protein (FosAKP) with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
1FTM
 
 | |
6TQB
 
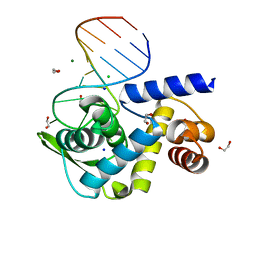 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE1 SL RNA motif | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
6NEU
 
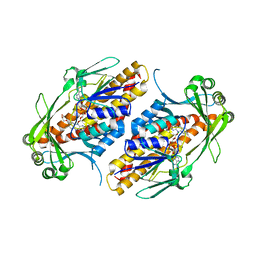 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
1YYW
 
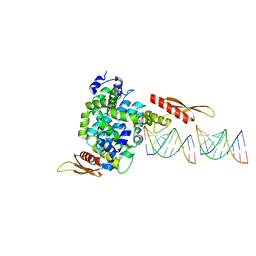 | | Crystal structure of RNase III from Aquifex aeolicus complexed with double stranded RNA at 2.8-Angstrom Resolution | | Descriptor: | 5'-R(*AP*AP*AP*UP*AP*UP*AP*UP*AP*UP*UP*U)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
2VB9
 
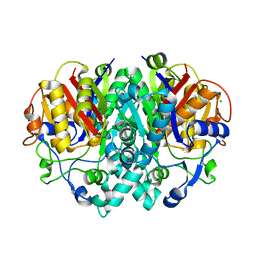 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli, apo structure | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, CHLORIDE ION | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4WS9
 
 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | Descriptor: | PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
6FRO
 
 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM potassium iodide. | | Descriptor: | IODIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6PSS
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5a) with TraR and mutant rpsT P2 promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6NY3
 
 | | CasX ternary complex with 30bp target DNA | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
