1P7K
 
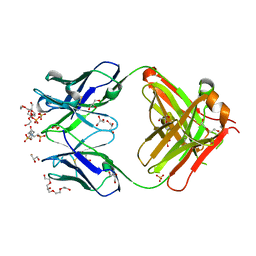 | | Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuermann, J.P, Henzl, M.T, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2003-05-02 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an anti-DNA fab complexed with a non-DNA ligand provides insights into cross-reactivity and molecular mimicry.
Proteins, 57, 2004
|
|
1P69
 
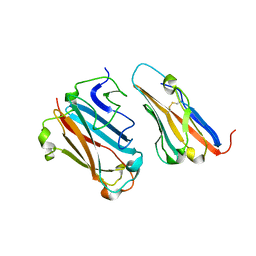 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (P417S MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1P6A
 
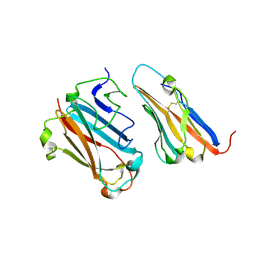 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1P4I
 
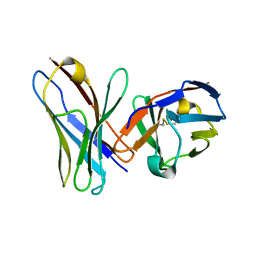 | | Crystal Structure of scFv against peptide GCN4 | | Descriptor: | ANTIBODY VARIABLE LIGHT CHAIN, antibody variable heavy chain | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1P4B
 
 | | Three-Dimensional Structure Of a Single Chain Fv Fragment Complexed With The peptide GCN4(7P-14P). | | Descriptor: | Antibody Variable heavy chain, Antibody Variable light chain, GCN4(7P-14P) peptide | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-22 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1OTU
 
 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTT
 
 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTS
 
 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1HKF
 
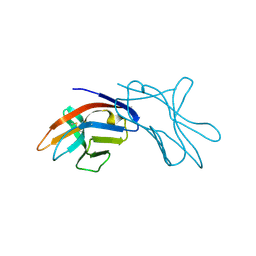 | | The three dimensional structure of NK cell receptor Nkp44, a triggering partner in natural cytotoxicity | | Descriptor: | NK CELL ACTIVATING RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-06-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Three-Dimensional Structure of the Human Nk Cell Receptor Nkp44, a Triggering Partner in Natural Cytotoxicity
Structure, 11, 2003
|
|
1OD9
 
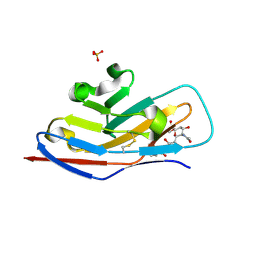 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-benzoyl-amino-9-deoxy-Neu5Ac (BENZ compound) | | Descriptor: | SIALOADHESIN, SULFATE ION, methyl 5-acetamido-3,5,9-trideoxy-9-[(phenylcarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1OD7
 
 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-(naphthyl-2-carbonyl)-amino-9-deoxy-Neu5Ac (NAP compound) | | Descriptor: | ME-A-9-N-(NAPHTHYL-2-CARBONYL)-AMINO-9-DEOXY-NEU5AC, SIALOADHESIN | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1ODA
 
 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-(biphenyl-4-carbonyl)-amino-9-deoxy-Neu5Ac (BIP compound) | | Descriptor: | ME-A-9-N-(BIPHENYL-4-CARBONYL)-AMINO-9-DEOXY-NEU5AC, SIALOADHESIN | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1OCW
 
 | |
1OB1
 
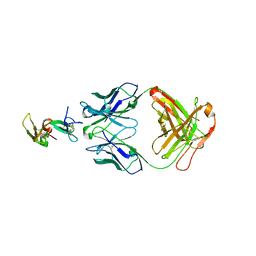 | | Crystal structure of a Fab complex whith Plasmodium falciparum MSP1-19 | | Descriptor: | ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pizarro, J.C, Chitarra, V, Verger, D, Holm, I, Petres, S, Dartville, S, Nato, F, Longacre, S, Bentley, G.A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Fab Complex Formed with Pfmsp1-19, the C-Terminal Fragment of Merozoite Surface Protein 1 from Plasmodium Falciparum: A Malaria Vaccine Candidate
J.Mol.Biol., 328, 2003
|
|
1OAZ
 
 | |
1OAX
 
 | |
1OAR
 
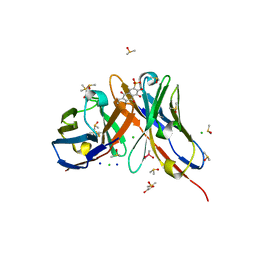 | | Fv IgE SPE-7 in complex with Alizarin Red | | Descriptor: | 1,2-ETHANEDIOL, ALIZARIN RED, CACODYLATE ION, ... | | Authors: | James, L.C, Roversi, P, Tawfik, D. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-15 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Antibody Multispecificity Mediated by Conformational Diversity
Science, 299, 2003
|
|
1OAU
 
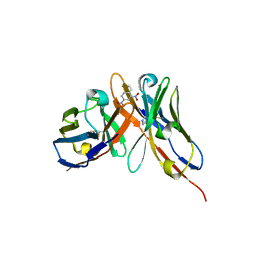 | | Fv Structure of the IgE SPE-7 in complex with DNP-Ser (immunising hapten) | | Descriptor: | 2,4-DINITROPHENOL, IMIDAZOLE, IMMUNOGLOBULIN E, ... | | Authors: | James, L.C, Roversi, P, Tawfik, D. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-15 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antibody Multispecificity Mediated by Conformational Diversity
Science, 299, 2003
|
|
1OAY
 
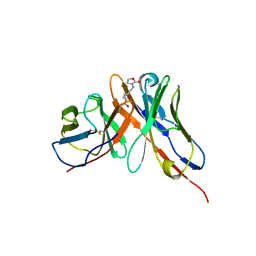 | |
1OAQ
 
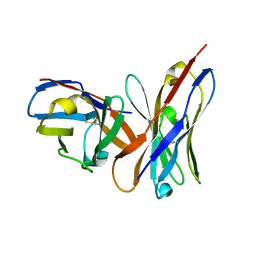 | |
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
1NN8
 
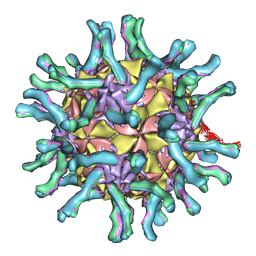 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J1X
 
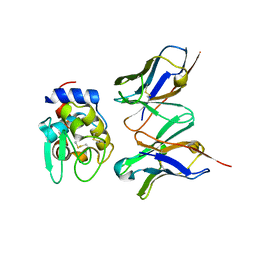 | | Crystal Structure of HyHEL-10 Fv mutant LS93A complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
1NGW
 
 | | Chimeric Affinity Matured Fab 7g12 complexed with mesoporphyrin | | Descriptor: | Mature Metal Chelatase Catalytic Antibody, Heavy chain, Light chain, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher IV, A.E, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-04 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
