1FDO
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
8GJ1
 
 | |
8GJ3
 
 | | E. coli clamp loader on primed template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in E. coli
To Be Published
|
|
8GJ2
 
 | |
8GJ0
 
 | |
6FAK
 
 | | Human afamin orthorhombic crystal form by controlled hydration | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rupp, B, Naschberger, A, Bowler, M.W. | | Deposit date: | 2017-12-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Controlled dehydration, structural flexibility and gadolinium MRI contrast compound binding in the human plasma glycoprotein afamin.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5NCO
 
 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | Descriptor: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2018-03-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
7X86
 
 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7XBN
 
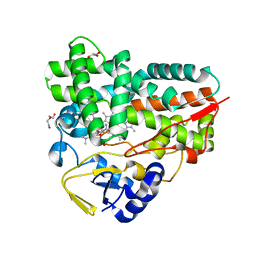 | | Crystal Structure of YC-17-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
5NPE
 
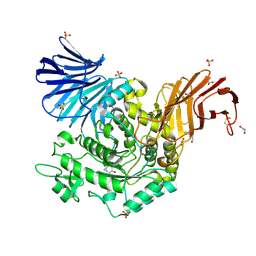 | | Crystal Structure of cjAgd31B (alpha-transglucosylase from Glycoside Hydrolase Family 31) in complex with beta Cyclophellitol Aziridine probe KY358 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5MZC
 
 | |
8DQI
 
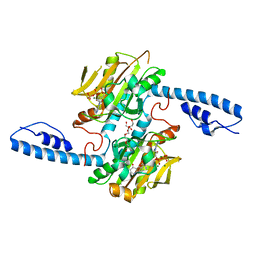 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 2- weeks of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
6D0P
 
 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8DQP
 
 | | Crystal structure of Arabidopsis thaliana COSY in complex with scopoletin | | Descriptor: | 1,2-ETHANEDIOL, 7-hydroxy-6-methoxy-2H-1-benzopyran-2-one, ACETATE ION, ... | | Authors: | Kim, C.Y, Mitchell, A.J, Gutierrez, M, Weng, J.K. | | Deposit date: | 2022-07-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Emergence of a proton exchange-based isomerization and lactonization mechanism in the plant coumarin synthase COSY.
Nat Commun, 14, 2023
|
|
5NGV
 
 | |
6FW3
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-007 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-(3,4-dimethoxyphenyl)-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)pyrrolidine-2,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-03-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-007
To be published
|
|
6CDA
 
 | | Crystal structure of L34A CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, GLYCEROL, ... | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
6CH0
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a glycerol molecule | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
2JXN
 
 | | Solution Structure of S. cerevisiae PDCD5-like Protein Ymr074cp | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Uncharacterized protein YMR074C | | Authors: | Hong, J, Zhang, J, Liu, Z, Shi, Y, Wu, J. | | Deposit date: | 2007-11-23 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of S. cerevisiae PDCD5-like Protein Ymr074cp Determined by Heteronuclear NMR Spectroscopy
To be Published
|
|
6G1Y
 
 | | Crystal structure of the photosensory core module (PCM) of a bathy phytochrome from Agrobacterium fabrum in the Pfr state. | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Schmidt, A, Qureshi, B.M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
1G8J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
6Q3R
 
 | | ASPERGILLUS ACULEATUS GALACTANASE | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, ... | | Authors: | Muderspach, S.J, Torpenholt, S, Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-12-04 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of Aspergillus aculeatus beta-1,4-galactanase in complex with galactobiose.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5LZ8
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 5-[2-[(4~{S})-4-~{tert}-butyl-2-oxidanylidene-pyrrolidin-1-yl]ethoxy]-2-fluoranyl-benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
8SVK
 
 | | Crystal structure of Bax D71N core domain BH3-groove dimer | | Descriptor: | Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Miller, M.S, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2023-05-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
