3GJ7
 
 | |
6Z1N
 
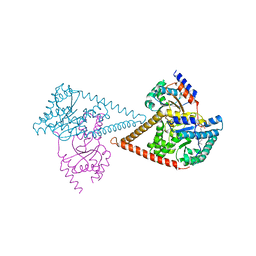 | | Structure of the human heterotetrameric cis-prenyltransferase complex | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, FARNESYL DIPHOSPHATE, ... | | Authors: | Lisnyansky Bar-El, M, Haitin, Y, Giladi, M. | | Deposit date: | 2020-05-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase complex.
Nat Commun, 11, 2020
|
|
7A94
 
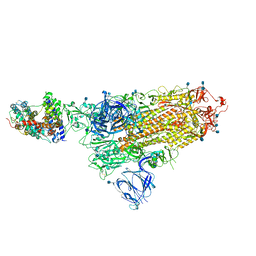 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
6ZNN
 
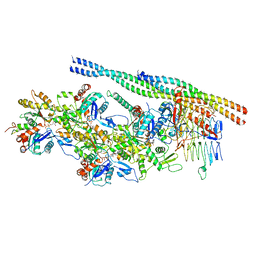 | | The pointed end complex of dynactin bound to Hook3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
4M63
 
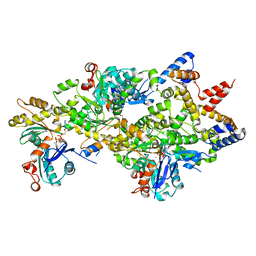 | | Crystal Structure of a Filament-Like Actin Trimer Bound to the Bacterial Effector VopL | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Zahm, J.A, Rosen, M.K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | The Bacterial Effector VopL Organizes Actin into Filament-like Structures.
Cell(Cambridge,Mass.), 155, 2013
|
|
3FTW
 
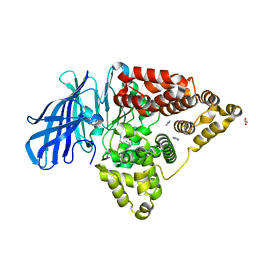 | |
6YSS
 
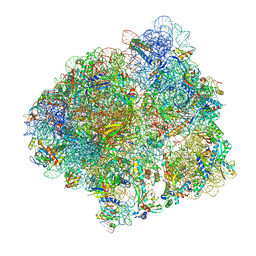 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
3FUD
 
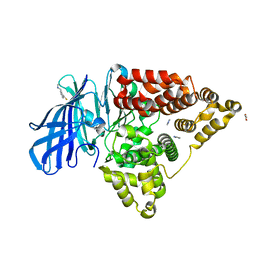 | |
6NTK
 
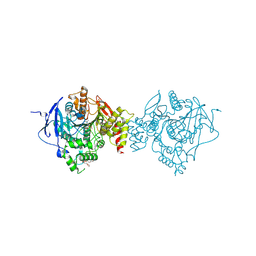 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-232 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
4GOO
 
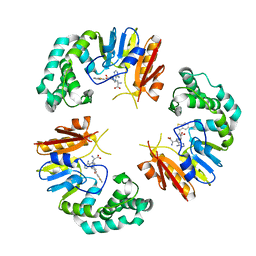 | |
6O2T
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ZP1
 
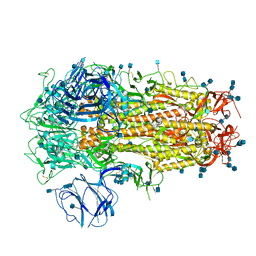 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6NUF
 
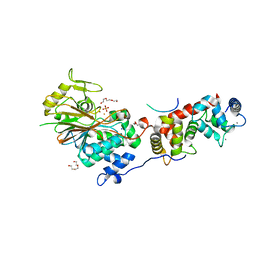 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
6ZHW
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 1 ps structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Branden, G, Neutze, R. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
4H0X
 
 | | Crystal structure of NAD+-Ia(E380A)-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-10 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6ZI6
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 20 ps structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
4GST
 
 | | REACTION COORDINATE MOTION IN AN SNAR REACTION CATALYZED BY GLUTATHIONE TRANSFERASE | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase.
Biochemistry, 32, 1993
|
|
6ZR2
 
 | | Cryo-EM structure of respiratory complex I in the active state from Mus musculus at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
6YHS
 
 | | Acinetobacter baumannii ribosome-amikacin complex - 50S subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
7BL6
 
 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL5
 
 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
4GWG
 
 | |
6NPH
 
 | | Structure of NKCC1 TM domain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
4GWT
 
 | | Structure of racemic Pin1 WW domain cocrystallized with DL-malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6NZ4
 
 | | YcjX-GDP (type I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
