7V7C
 
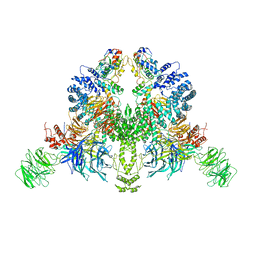 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | Authors: | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
3MEE
 
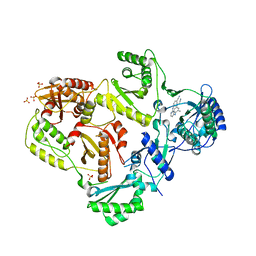 | | HIV-1 Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
1PTG
 
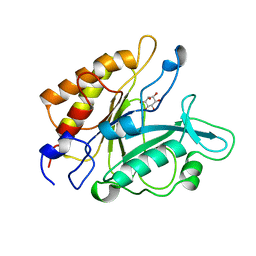 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C IN COMPLEX WITH MYO-INOSITOL | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W, Ryan, M, Bullock, T.L, Griffith, O.H. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol.
EMBO J., 14, 1995
|
|
4BC4
 
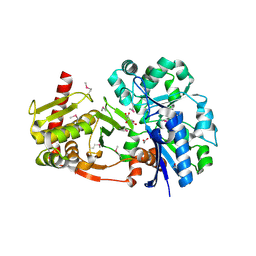 | | Crystal structure of human D-xylulokinase in complex with D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, D-XYLULOSE, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
6SZQ
 
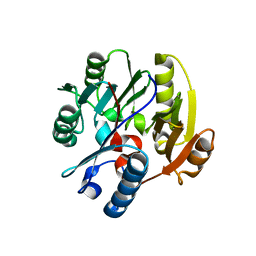 | | Crystal structure of human DDAH-1 | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Hennig, S, Vetter, I.R, Schade, D. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
J.Med.Chem., 63, 2020
|
|
5I78
 
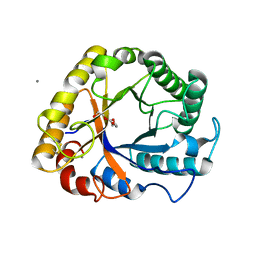 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5JE5
 
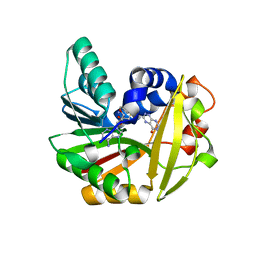 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and 1-demethyltoxoflavin | | Descriptor: | 6-methylpyrimido[5,4-e][1,2,4]triazine-5,7(6H,8H)-dione, Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
6I8S
 
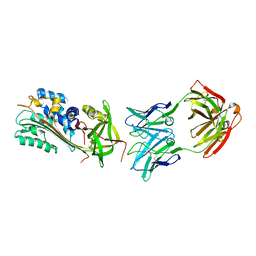 | |
7UF4
 
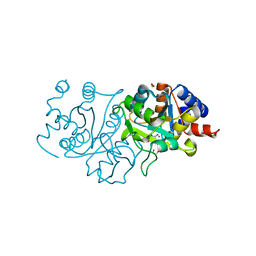 | | RibB from Vibrio cholera bound with intermediate 1 of the reaction cycle and D-ribulose-5-phosphate (D-Ru5P) | | Descriptor: | (2R)-2-hydroxy-3,4-dioxopentyl dihydrogen phosphate, 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, ... | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
5JHQ
 
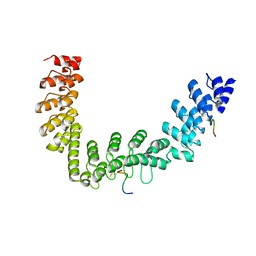 | |
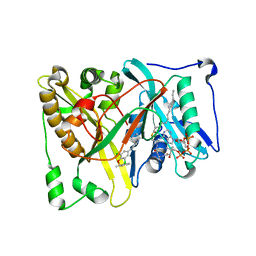
5T6C
 
 | |
6BVU
 
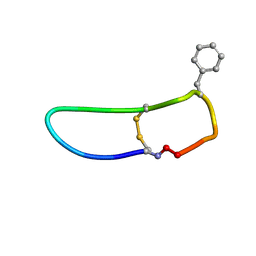 | | SFTI-HFRW-1 | | Descriptor: | Trypsin inhibitor 1 HFRW-1 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
4NF7
 
 | |
3MEG
 
 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
5K4P
 
 | | Catalytic Domain of MCR-1 phosphoethanolamine transferase | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, sorbitol | | Authors: | Stojanoski, V, Palzkill, T, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2016-05-21 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Structure of the catalytic domain of the colistin resistance enzyme MCR-1.
Bmc Biol., 14, 2016
|
|
6SBJ
 
 | | X-ray structure of mus musculus Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) apo-form uuncomplexed | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
6SW5
 
 | | Crystal structure of the human S-adenosylmethionine synthetase 1 (ligand-free form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, S-adenosylmethionine synthase isoform type-1 | | Authors: | Panmanee, J, Antoyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the dominant inheritance of hypermethioninemia associated with the Arg264His mutation in the MAT1A gene.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6FCD
 
 | | Human Methionine Adenosyltransferase II mutant (R264A) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
5JGT
 
 | | Human carbonic anhydrase II (F131Y/L198A) complexed with 1,3-thiazole-2-sulfonamide | | Descriptor: | 1,3-thiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5GK4
 
 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 2.0 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 2.0 Angstrom resolution
To Be Published
|
|
6UB2
 
 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Endo-beta-1,3-glucanase, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
1M2P
 
 | | Crystal structure of 1,8-di-hydroxy-4-nitro-anthraquinone/CK2 kinase complex | | Descriptor: | 1,8-DI-HYDROXY-4-NITRO-ANTHRAQUINONE, Casein kinase II, alpha chain | | Authors: | De Moliner, E, Moro, S, Sarno, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
8XI6
 
 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 98, 2024
|
|
8J6S
 
 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
