8A1B
 
 | |
2BEK
 
 | |
6NUE
 
 | | Small conformation of apo CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
1HSN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
7NYY
 
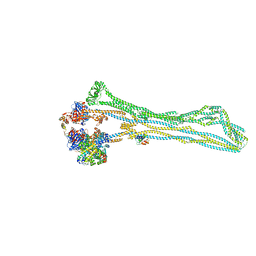 | | Cryo-EM structure of the MukBEF monomer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
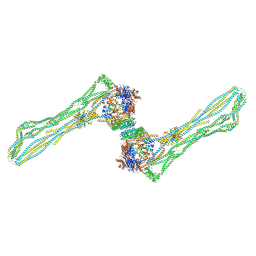 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
1KEJ
 
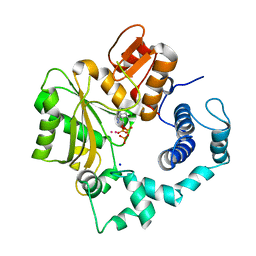 | | Crystal Structure of Murine Terminal Deoxynucleotidyl Transferase complexed with ddATP | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
7Z4D
 
 | |
1PO6
 
 | |
7NPD
 
 | | Vibiro cholerae ParA2 | | Descriptor: | Walker A-type ATPase | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
6OAB
 
 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6NUD
 
 | | Small conformation of ssRNA-bound CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
2AQF
 
 | | Structural and functional analysis of ADA2 alpha swirm domain | | Descriptor: | transcriptional adaptor 2, Ada2 alpha | | Authors: | Qian, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and chromosomal DNA binding of the SWIRM domain.
Nat.Struct.Mol.Biol., 12, 2005
|
|
6TX3
 
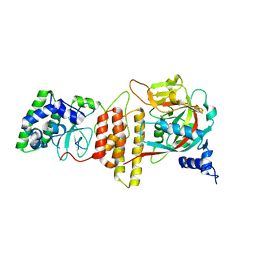 | | HPF1 bound to catalytic fragment of PARP2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2,Poly [ADP-ribose] polymerase 2 | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
2AQE
 
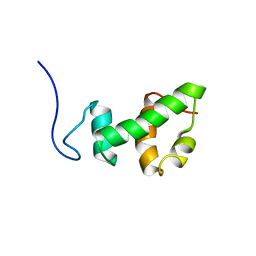 | | Structural and functional analysis of ada2 alpha swirm domain | | Descriptor: | Transcriptional adaptor 2, Ada2 alpha | | Authors: | Qian, C, Zhang, Q, Zeng, L, Zhou, M.-M. | | Deposit date: | 2005-08-17 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and chromosomal DNA binding of the SWIRM domain
Nat.Struct.Mol.Biol., 12, 2005
|
|
4DT1
 
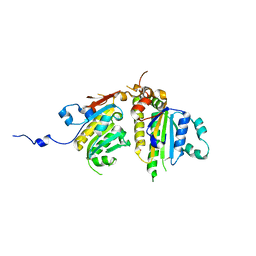 | | Crystal structure of the Psy3-Csm2 complex | | Descriptor: | Chromosome segregation in meiosis protein 2, ETHANOL, Platinum sensitivity protein 3 | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-20 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage
J.Biol.Chem., 287, 2012
|
|
4DAV
 
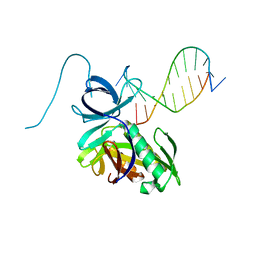 | | The structure of Pyrococcus Furiosus SfsA in complex with DNA | | Descriptor: | 5'-D(*CP*GP*CP*TP*GP*TP*CP*TP*CP*GP*CP*T)-3', Sugar fermentation stimulation protein homolog | | Authors: | Baker, P.J, Allen, F.L. | | Deposit date: | 2012-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of SfsA and its DNA complex; A DNA/RNA nuclease with a novel domain combination
To be Published
|
|
3BVQ
 
 | | Crystal Structure of Apo NotI Restriction Endonuclease | | Descriptor: | FE (III) ION, NotI restriction endonuclease, SULFATE ION | | Authors: | Lambert, A.R, Sussman, D, Shen, B, Stoddard, B.L. | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-22 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Rare-Cutting Restriction Endonuclease NotI Reveal a Unique Metal Binding Fold Involved in DNA Binding.
Structure, 16, 2008
|
|
5AHO
 
 | | Crystal structure of human 5' exonuclease Apollo | | Descriptor: | 1,2-ETHANEDIOL, 5' EXONUCLEASE APOLLO, L(+)-TARTARIC ACID, ... | | Authors: | Allerston, C.K, Vollmar, M, Krojer, T, Pike, A.C.W, Newman, J.A, Carpenter, E, Quigley, A, Mahajan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Gileadi, O. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
5B2T
 
 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
1N4B
 
 | | Solution Structure of the undecamer CGAAAC*TTTCG | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*D00*TP*TP*TP*CP*G)-3' | | Authors: | Webba da Silva, M, Noronha, A.M, Noll, D.M, Miller, P.S, Colvin, O.M, Gamcsik, M.P. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing
Mispair-Aligned N4C-Ethyl-N4C
Interstrand Cross-Linked Cytosines
Biochemistry, 41, 2002
|
|
6IPU
 
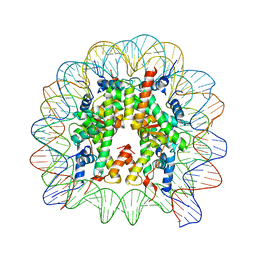 | |
1EFY
 
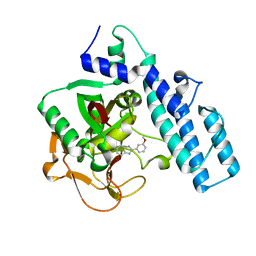 | | CRYSTAL STRUCTURE OF THE CATALYTIC FRAGMENT OF POLY (ADP-RIBOSE) POLYMERASE COMPLEXED WITH A BENZIMIDAZOLE INHIBITOR | | Descriptor: | 2-(3'-METHOXYPHENYL) BENZIMIDAZOLE-4-CARBOXAMIDE, POLY (ADP-RIBOSE) POLYMERASE | | Authors: | White, A.W, Almassy, R, Calvert, A.H, Curtin, N.J, Griffin, R.J, Hostomsky, Z, Maegley, K, Newell, D.R, Srinivasan, S, Golding, B.T. | | Deposit date: | 2000-02-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase.
J.Med.Chem., 43, 2000
|
|
6CC2
 
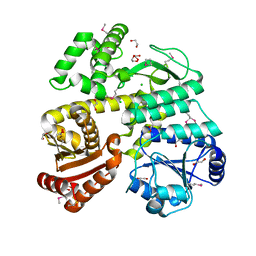 | | Crystal Structure of CDC45 from Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cell division control protein 45 cdc45 putative, ... | | Authors: | Shi, K, Kurniawan, F, Kurahashi, K, Bielinsky, A, Aihara, H. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure ofEntamoeba histolyticaCdc45 Suggests a Conformational Switch that May Regulate DNA Replication.
iScience, 3, 2018
|
|
5DA9
 
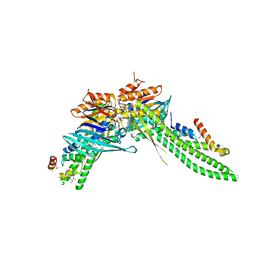 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with the Rad50-binding domain of Mre11 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative double-strand break protein, ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
