6D0Q
 
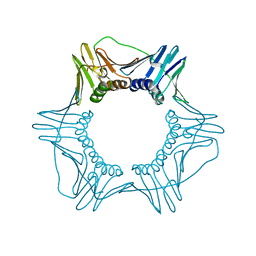 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.80051017 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
4FTG
 
 | |
6D0R
 
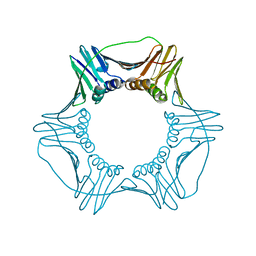 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85856962 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
6CZQ
 
 | |
6CIX
 
 | |
6D6R
 
 | |
1O9G
 
 | |
1PK2
 
 | |
3RPG
 
 | | Bmi1/Ring1b-UbcH5c complex structure | | Descriptor: | E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | Bentley, M.L, Dong, K.C, Cochran, A.G. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6485 Å) | | Cite: | Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex.
Embo J., 30, 2011
|
|
6L53
 
 | | Structure of SMG1 | | Descriptor: | Serine/threonine-protein kinase SMG1 | | Authors: | Xu, Y, Qi, Y. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structure of SMG1-SMG8-SMG9 complex.
Cell Res., 29, 2019
|
|
3HV4
 
 | | Human p38 MAP Kinase in Complex with RL51 | | Descriptor: | 1-{3-[(6-aminoquinolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
3HL7
 
 | | Crystal Structure of Human p38alpha complexed with SD-0006 | | Descriptor: | 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, 2-{4-[5-(4-chlorophenyl)-4-pyrimidin-4-yl-1H-pyrazol-3-yl]piperidin-1-yl}-2-oxoethanol, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Kurumbail, R.G, Stegeman, R.A, Williams, J.M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural bioinformatics-based prediction of exceptional selectivity of p38 MAP kinase inhibitor PH-797804.
Biochemistry, 48, 2009
|
|
3HLL
 
 | | Crystal Structure of Human p38alpha complexed with PH-797804 | | Descriptor: | 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, 3-{3-bromo-4-[(2,4-difluorobenzyl)oxy]-6-methyl-2-oxopyridin-1(2H)-yl}-N,4-dimethylbenzamide, HYPOPHOSPHITE, ... | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Kurumbail, R.G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural bioinformatics-based prediction of exceptional selectivity of p38 MAP kinase inhibitor PH-797804.
Biochemistry, 48, 2009
|
|
3HL8
 
 | | Crystal structure of exonuclease I in complex with inhibitor BCBP | | Descriptor: | (5R)-3-tert-butyl-1-(6-chloro-1,3-benzothiazol-2-yl)-4,5-dihydro-1H-pyrazol-5-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Satyshur, K.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1WBT
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-5-MORPHOLIN-4-YL-N-[1-(2-PYRIDIN-4-YLETHYL)-1H-INDOL-6-YL]BENZAMIDE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
1WC8
 
 | | The crystal structure of mouse bet3p | | Descriptor: | MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | Authors: | Kim, Y.-G, Sacher, M, Oh, B.-H. | | Deposit date: | 2004-11-10 | | Release date: | 2004-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
3HP9
 
 | | Crystal structure of SSB/Exonuclease I in complex with inhibitor CFAM | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-chloro-5-(trifluoromethyl)phenyl]amino}-5-methoxybenzoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Satyshur, K.A. | | Deposit date: | 2009-06-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1WI4
 
 | | Solution structure of the PDZ domain of syntaxin binding protein 4 | | Descriptor: | syntaxin binding protein 4 | | Authors: | Endo, H, Tomizawa, T, Kigawa, T, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of syntaxin binding protein 4
To be published
|
|
3HUB
 
 | | Human p38 MAP Kinase in Complex with Scios-469 | | Descriptor: | 2-(6-chloro-5-{[(2R,5S)-4-(4-fluorobenzyl)-2,5-dimethylpiperazin-1-yl]carbonyl}-1-methyl-1H-indol-3-yl)-N,N-dimethyl-2-oxoacetamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-06-13 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fluorophore labeling of the glycine-rich loop as a method of identifying inhibitors that bind to active and inactive kinase conformations.
J.Am.Chem.Soc., 132, 2010
|
|
6DLN
 
 | |
3HUF
 
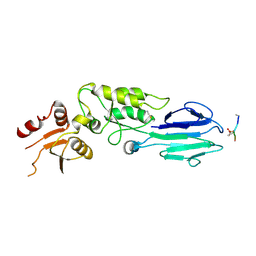 | | Structure of the S. pombe Nbs1-Ctp1 complex | | Descriptor: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | Authors: | Williams, R.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-06-13 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
3HSU
 
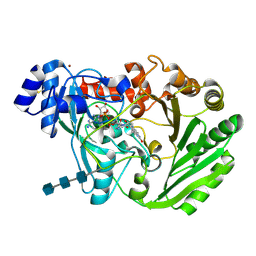 | | Functional roles of the 6-s-cysteinyl, 8 alpha-N1-histidyl FAD in Glucooligosaccharide Oxidase from Acremonium strictum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Liaw, S.-H, Huang, C.-H. | | Deposit date: | 2009-06-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Functional roles of the 6-S-cysteinyl, 8alpha-N1-histidyl FAD in glucooligosaccharide oxidase from Acremonium strictum
J.Biol.Chem., 283, 2008
|
|
3HTX
 
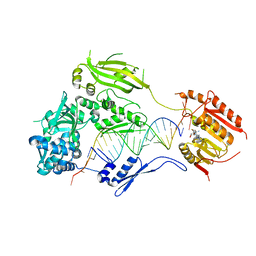 | | Crystal structure of small RNA methyltransferase HEN1 | | Descriptor: | 5'-R(*GP*AP*UP*UP*UP*CP*UP*CP*UP*CP*UP*GP*CP*AP*AP*GP*CP*GP*AP*AP*AP*G)-3', 5'-R(P*UP*UP*CP*GP*CP*UP*UP*GP*CP*AP*GP*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*C)-3', HEN1, ... | | Authors: | Huang, Y, Ji, L.-J, Huang, Q.-C, Vassylyev, D.G, Chen, X.-M, Ma, J.-B. | | Deposit date: | 2009-06-12 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into mechanisms of the small RNA methyltransferase HEN1.
Nature, 461, 2009
|
|
1WKQ
 
 | |
1WC9
 
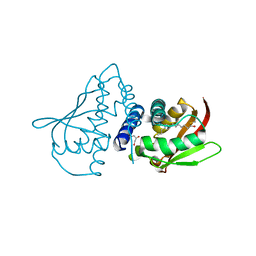 | | The crystal structure of truncated mouse bet3p | | Descriptor: | GLYCEROL, MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | Authors: | Kim, Y.-G, Lee, H.-S, Sacher, M, Oh, B.-H. | | Deposit date: | 2004-11-10 | | Release date: | 2004-12-13 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
