2RH4
 
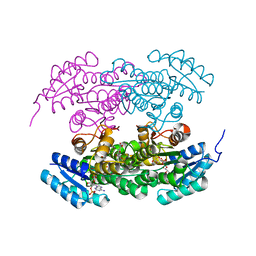 | | Actinorhodin ketoreductase, actKR, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2RHC
 
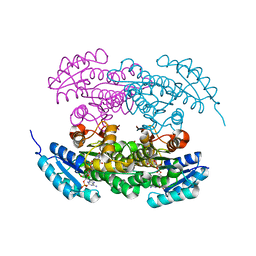 | | Actinorhodin ketordeuctase, actKR, with NADP+ and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-08 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
5WNM
 
 | |
5WNL
 
 | |
3LIM
 
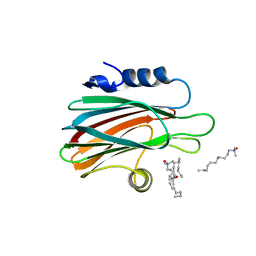 | | Crystal structure of the pore forming toxin frac from sea anemone actinia fragacea | | Descriptor: | Fragaceatoxin C, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Mechaly, A.E, Bellomio, A, Morante, K, Gonzalez-Manas, J.M, Guerin, D.M.A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins.
Structure, 19, 2011
|
|
6S1F
 
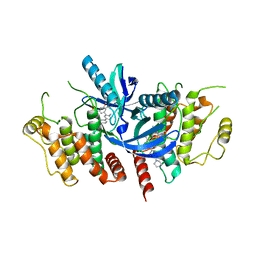 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[3-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-5-methoxy-phenyl]methanesulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
To be published
|
|
4QAM
 
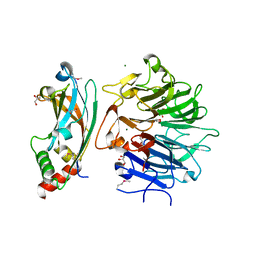 | | Crystal Structure of the RPGR RCC1-like domain in complex with the RPGR-interacting domain of RPGRIP1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, X-linked retinitis pigmentosa GTPase regulator, ... | | Authors: | Remans, K, Buerger, M, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | C2 domains as protein-protein interaction modules in the ciliary transition zone.
Cell Rep, 8, 2014
|
|
4H6V
 
 | |
7R1V
 
 | |
7R1W
 
 | | E. coli BAM complex (BamABCDE) bound to dynobactin A | | Descriptor: | Dynobactin A, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Hiller, S, Maier, T. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
3HO4
 
 | |
3HO3
 
 | |
6R5F
 
 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
6C95
 
 | |
3RGI
 
 | |
3SBM
 
 | | Trans-acting transferase from Disorazole synthase in complex with Acetate | | Descriptor: | ACETATE ION, DisD protein, HEXAETHYLENE GLYCOL | | Authors: | Khosla, C, Mathews, I.I, Wong, F.T, Jin, X. | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Mechanism of the trans-Acting Acyltransferase from the Disorazole Synthase.
Biochemistry, 50, 2011
|
|
4TSO
 
 | | Crystal structure of FraC with DHPC bound (crystal form I) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSP
 
 | | Crystal structure of FraC with DHPC bound (crystal form II) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSL
 
 | | Crystal structure of FraC with POC bound (crystal form I) | | Descriptor: | ACETATE ION, FORMIC ACID, Fragaceatoxin C, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSY
 
 | | Crystal structure of FraC with lipids | | Descriptor: | 2-[[(E,2S,3R)-2-(hexanoylamino)-3-oxidanyl-dec-4-enoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, Fragaceatoxin C, HEPTANE-1,2,3-TRIOL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSN
 
 | | Crystal structure of FraC with POC bound (crystal form II) | | Descriptor: | ACETATE ION, Fragaceatoxin C, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSQ
 
 | | Crystal structure of FraC with DHPC bound (crystal form III) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, Fragaceatoxin C, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
2GZS
 
 | |
2GZR
 
 | |
7MDE
 
 | | Full-length S95A ClbP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Velilla, J.A, Volpe, M.R, Gaudet, R. | | Deposit date: | 2021-04-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of colibactin activation by the ClbP peptidase.
Nat.Chem.Biol., 19, 2023
|
|
