3UEQ
 
 | | Crystal structure of amylosucrase from Neisseria polysaccharea in complex with turanose | | Descriptor: | 3-O-alpha-D-glucopyranosyl-D-fructose, Amylosucrase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guerin, F, Pizzut-Serin, S, Potocki-Veronese, G, Guillet, V, Mourey, L, Remaud-Simeon, M, Andre, I, Tranier, S. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Investigation of the Thermostability and Product Specificity of Amylosucrase from the Bacterium Deinococcus geothermalis.
J.Biol.Chem., 287, 2012
|
|
3HHY
 
 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with catechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CATECHOL, CHLORIDE ION, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
1J7H
 
 | | Solution Structure of HI0719, a Hypothetical Protein From Haemophilus Influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI0719 | | Authors: | Parsons, L, Bonander, N, Eisenstein, E, Gilson, M, Kairys, V, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Ligand Screening of HI0719, a Highly Conserved Protein from Bacteria to Humans in the YjgF/YER057c/UK114 Family
Biochemistry, 42, 2003
|
|
1AYG
 
 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1B01
 
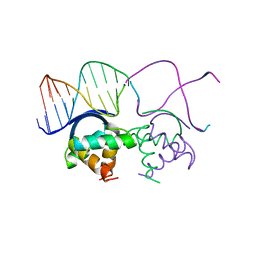 | | TRANSCRIPTIONAL REPRESSOR COPG/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*GP*CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*GP*CP*AP*TP*TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*G)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
3I4V
 
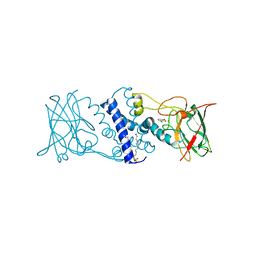 | | Crystal structure determination of catechol 1,2-dioxygenase from rhodococcus opacus 1CP in complex with 3-chlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3-chlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3I51
 
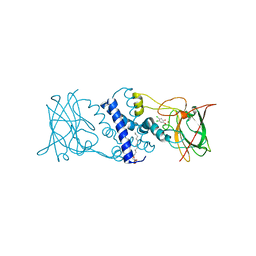 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with 4,5-dichlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 4,5-dichlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3HUV
 
 | |
5CGQ
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with F9 ligand in the alpha-site and the product L-Tryptophan in the beta-site. | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2015-07-09 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with F9 ligand and the product L-Tryptophan in the beta-site.
To Be Published
|
|
6ATB
 
 | | Crystal Structure of human NAMPT in complex with NVP-LOD812 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-N'-[(pyridin-3-yl)methyl]urea, ... | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-08-28 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
3LY0
 
 | | Crystal structure of metallo peptidase from Rhodobacter sphaeroides liganded with phosphinate mimic of dipeptide L-Ala-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Dipeptidase AC. Metallo peptidase. MEROPS family M19, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of metallo peptidase from Rhodobacter sphaeroides
liganded with phosphinate mimic of dipeptide L-Ala-D-Ala
To be Published
|
|
5C5G
 
 | |
4F03
 
 | |
1BEI
 
 | | Shk-dnp22: A Potent Kv1.3-specific immunosuppressive polypeptide, NMR, 20 structures | | Descriptor: | POTASSIUM CHANNEL TOXIN SHK | | Authors: | Kalman, K, Pennington, M.W, Lanigan, M.D, Nguyen, A, Rauer, H, Mahnir, V, Gutman, G.A, Paschetto, K, Kem, W.R, Grissmer, S, Christian, E.P, Cahalan, M.D, Norton, R.S, Chandy, K.G. | | Deposit date: | 1998-05-14 | | Release date: | 1998-12-02 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide.
J.Biol.Chem., 273, 1998
|
|
3IK5
 
 | |
3QK8
 
 | |
1UFG
 
 | | Solution structure of immunoglobulin like domain of mouse nuclear lamin | | Descriptor: | Lamin A | | Authors: | Kobayashi, N, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-29 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of immunoglobulin like domain of mouse nuclear lamin
To be Published
|
|
3MHG
 
 | | Dihydroxyacetone phosphate carbanion intermediate in tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, CALCIUM ION, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | LowKam, C, Liotard, B. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-28 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a class I tagatose-1,6-bisphosphate aldolase: investigation into an apparent loss of stereospecificity.
J.Biol.Chem., 285, 2010
|
|
4CC3
 
 | | Complex of human Tuba C-terminal SH3 domain and Mena proline-rich peptide - H3 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, DYNAMIN-BINDING PROTEIN, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
2RH8
 
 | | Structure of apo anthocyanidin reductase from vitis vinifera | | Descriptor: | Anthocyanidin reductase, CHLORIDE ION | | Authors: | Gargouri, M, Mauge, C, Langlois D'Estaintot, B, Granier, T, Manigan, C, Gallois, B. | | Deposit date: | 2007-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4C6G
 
 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
2RPZ
 
 | | Solution structure of the monomeric form of mouse APOBEC2 | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Hayashi, F, Nagata, T, Nagashima, T, Muto, Y, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monomeric form of mouse APOBEC2
To be Published
|
|
3CHF
 
 | |
1UEQ
 
 | | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein) | | Descriptor: | MEMBRANE ASSOCIATED GUANYLATE KINASE INVERTED-2 (MAGI-2) | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein)
To be Published
|
|
4CC2
 
 | | Complex of human Tuba C-terminal SH3 domain with human N-WASP proline- rich peptide - P212121 | | Descriptor: | CHLORIDE ION, DYNAMIN-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
