4P1L
 
 | | Crystal structure of a trap periplasmic solute binding protein from chromohalobacter salexigens dsm 3043 (csal_2479), target EFI-510085, with bound d-glucuronate, spg i213 | | Descriptor: | GLYCEROL, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
195D
 
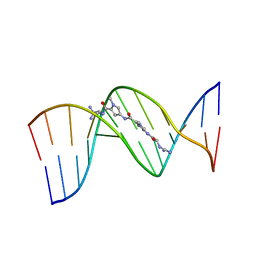 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
3NYE
 
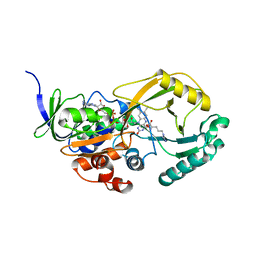 | |
1OJ8
 
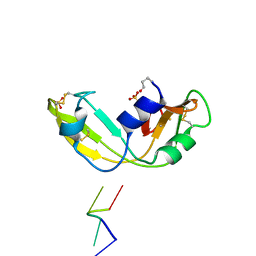 | | Novel and retro Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with d(ApCpGpA) and (2',5'CpG) | | Descriptor: | 5'-D(*AP*CP*GP*AP)-3', RC-RNASE6 RIBONUCLEASE, SULFATE ION | | Authors: | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-Y, Cheng, P.-T, Sun, Y.-J. | | Deposit date: | 2003-07-07 | | Release date: | 2004-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retro and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with (2',5'Cpg) and D(Apcpgpa)
To be Published
|
|
3WQD
 
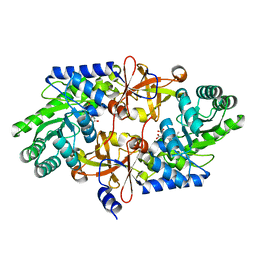 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-erythro-3-hydroxyaspartate | | Descriptor: | (3S)-3-hydroxy-D-aspartic acid, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
110D
 
 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*GP*CP*CP*G)-3') | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1UB8
 
 | | Crystal structure of d(GCGAAGC), bending duplex with a bulge-in residue | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*GP*C)-3', COBALT HEXAMMINE(III) | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-03-31 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1K55
 
 | | OXA 10 class D beta-lactamase at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Beta lactamase OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1XAS
 
 | | CRYSTAL STRUCTURE, AT 2.6 ANGSTROMS RESOLUTION, OF THE STREPTOMYCES LIVIDANS XYLANASE A, A MEMBER OF THE F FAMILY OF BETA-1,4-D-GLYCANSES | | Descriptor: | 1,4-BETA-D-XYLAN XYLANOHYDROLASE | | Authors: | Derewenda, U, Derewenda, Z.S. | | Deposit date: | 1994-05-31 | | Release date: | 1995-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure, at 2.6-A resolution, of the Streptomyces lividans xylanase A, a member of the F family of beta-1,4-D-glycanases.
J.Biol.Chem., 269, 1994
|
|
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
2LS8
 
 | | Solution structure of human C-type lectin domain family 4 member D | | Descriptor: | C-type lectin domain family 4 member D | | Authors: | Harris, R, Gaudette, J, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human C-type lectin domain family 4 member D
To be Published
|
|
2BES
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronohydroxamic acid. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism.
J.Biol.Chem., 280, 2005
|
|
2XPV
 
 | | TetR(D) in complex with minocycline and magnesium. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dalm, D, Proft, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Tetr(D) in Complex with Minocycline.
To be Published
|
|
4DNS
 
 | | Crystal structure of Bermuda grass isoallergen BG60 provides insight into the various cross-allergenicity of the pollen group 4 allergens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FAD-linked oxidoreductase BG60, ... | | Authors: | Huang, T.H, Peng, H.J, Su, S.N, Liaw, S.H. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Various cross-reactivity of the grass pollen group 4 allergens: crystallographic study of the Bermuda grass isoallergen Cyn d 4.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1H76
 
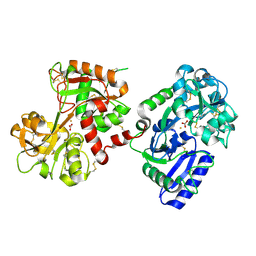 | | The crystal structure of diferric porcine serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-03 | | Release date: | 2002-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal and Molecular Structures of Diferric Porcine and Rabbit Serum Transferrins at Resolutions of 2.15 And 2.60A, Respectively
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2DWU
 
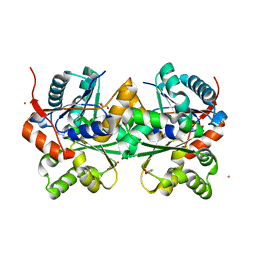 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
1L5R
 
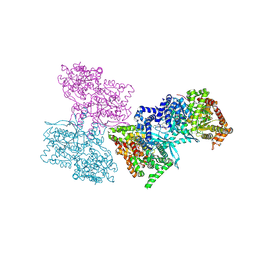 | | Human liver glycogen phosphorylase a complexed with riboflavin, N-Acetyl-beta-D-Glucopyranosylamine and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, N-acetyl-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
1LMP
 
 | |
2XPW
 
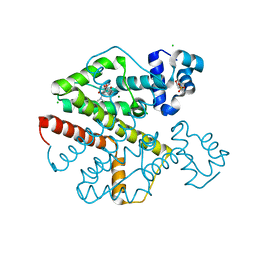 | | TetR(D) in complex with oxytetracycline and magnesium. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dalm, D, Proft, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Tetr(D) in Complex with Oxytetracycline.
To be Published
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
2BET
 
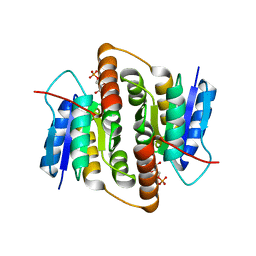 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronate. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONATE, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism
J.Biol.Chem., 280, 2005
|
|
1L5S
 
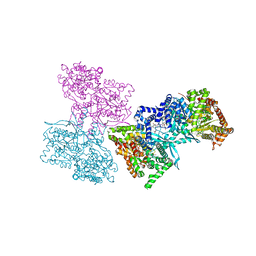 | | Human liver glycogen phosphorylase complexed with uric acid, N-Acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
1L7X
 
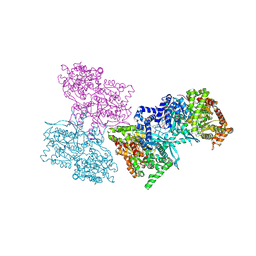 | | Human liver glycogen phosphorylase b complexed with caffeine, N-acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CAFFEINE, Glycogen phosphorylase, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-18 | | Release date: | 2002-12-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
4N8G
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0660), Target EFI-501075, with bound D-alanine-D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
