3CP6
 
 | | Crystal structure of human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor | | Descriptor: | (4aS,7aR)-octahydro-1H-cyclopenta[b]pyridine-6,6-diylbis(phosphonic acid), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, von Delft, F, Barnett, B.L, Ebetino, F.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor.
To be Published
|
|
4LKO
 
 | | Crystal structure of human DPP-IV in complex with BMS-744891 | | Descriptor: | 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-6,7-dihydro-5H-pyrrolo[3,4-b]pyridin-5-one, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-07-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
3K38
 
 | |
3K3A
 
 | |
3K14
 
 | | Co-crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 535, ethyl 3-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole-2-carboxylate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
3CO9
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
9BJA
 
 | |
4URL
 
 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URN
 
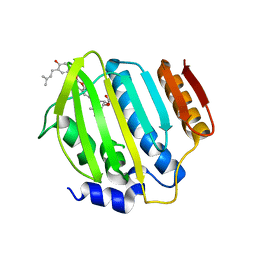 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4W2O
 
 | |
5X4M
 
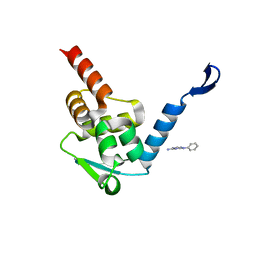 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
8REF
 
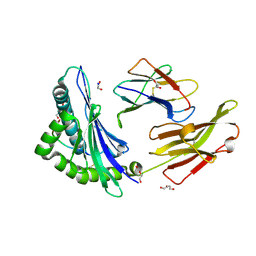 | | Crystal structure of HLA B*13:01 in complex with SVLNDILARL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RCV
 
 | | Crystal structure of HLA B*13:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen B alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
9N2C
 
 | | Impacts of ribosomal RNA sequence variation on gene expression and phenotype: Cryo-EM structure of the rrsH ribosome (HBB-70S) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA (rRNA) from the rrnH operon, 23S ribosomal RNA (rRNA) from the rrnB operon, ... | | Authors: | Welfer, G.A, Brady, R.A, Natchiar, S.K, Watson, Z.L, Rundlet, E.J, Alejo, J.L, Singh, A.P, Mishra, N.K, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2025-01-28 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Impacts of ribosomal RNA sequence variation on gene expression and phenotype.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 380, 2025
|
|
3S7A
 
 | | Human dihydrofolate reductase binary complex with PT684 | | Descriptor: | 4-({5-[(2,4-diaminopteridin-6-yl)methyl]-5H-dibenzo[b,f]azepin-2-yl}oxy)butanoic acid, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Pace, J, Nowak, J. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of human dihydrofolate reductase as a binary complex with the potent and selective inhibitor 2,4-diamino-6-{2'-O-(3-carboxypropyl)oxydibenz[b,f]-azepin-5-yl}methylpteridine reveals an unusual binding mode.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1KMR
 
 | |
1LFH
 
 | | MOLECULAR REPLACEMENT SOLUTION OF THE STRUCTURE OF APOLACTOFERRIN, A PROTEIN DISPLAYING LARGE-SCALE CONFORMATIONAL CHANGE | | Descriptor: | CHLORIDE ION, LACTOFERRIN | | Authors: | Anderson, B.F, Baker, E.N, Norris, G.E. | | Deposit date: | 1991-09-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular replacement solution of the structure of apolactoferrin, a protein displaying large-scale conformational change.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
9L5R
 
 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
3SEB
 
 | | STAPHYLOCOCCAL ENTEROTOXIN B | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN B | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1997-11-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of microbial superantigen staphylococcal enterotoxin B at 1.5 A resolution: implications for superantigen recognition by MHC class II molecules and T-cell receptors.
J.Mol.Biol., 277, 1998
|
|
9BE0
 
 | | GC-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6TG6
 
 | | Toprim domain of RNase M5 | | Descriptor: | Ribonuclease M5 | | Authors: | Oerum, S, Catala, M, Tisne, C. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
9BDZ
 
 | | TA-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, ZINC ION, double-stranded DNA | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2VKS
 
 | |
1TXA
 
 | | SOLUTION NMR STRUCTURE OF TOXIN B, A LONG NEUROTOXIN FROM THE VENOM OF THE KING COBRA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TOXIN B | | Authors: | Peng, S.-S, Kumar, T.K.S, Jayaraman, G, Chang, C.-C, Yu, C. | | Deposit date: | 1996-07-20 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of toxin b, a long neurotoxin from the venom of the king cobra (Ophiophagus hannah).
J.Biol.Chem., 272, 1997
|
|
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
