1D62
 
 | |
1D90
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH I.T MISMATCHED BASE PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*IP*GP*CP*TP*CP*C)-3') | | Authors: | Cruse, W.B.T, Aymani, J, Kennard, O, Brown, T, Jack, A.G.C, Leonard, G.A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs.
Nucleic Acids Res., 17, 1989
|
|
2PCB
 
 | |
1XLA
 
 | |
2AVI
 
 | |
1XLE
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1D92
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH G.T MISMATCHED BASE-PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*TP*CP*C)-3') | | Authors: | Hunter, W.N, Kneale, G, Brown, T, Rabinovich, D, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs.
J.Mol.Biol., 190, 1986
|
|
1CTY
 
 | |
1XLK
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XIN
 
 | |
1XLG
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, MAGNESIUM ION, ... | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLJ
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLH
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLL
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, ZINC ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLI
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | 5-thio-alpha-D-glucopyranose, D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
3RAT
 
 | |
2ABD
 
 | |
2CRD
 
 | | ANALYSIS OF SIDE-CHAIN ORGANIZATION ON A REFINED MODEL OF CHARYBDOTOXIN: STRUCTURAL AND FUNCTIONAL IMPLICATIONS | | Descriptor: | CHARYBDOTOXIN | | Authors: | Bontems, F, Roumestand, C, Gilquin, B, Menez, A, Toma, F. | | Deposit date: | 1993-02-17 | | Release date: | 1993-07-15 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications.
Biochemistry, 31, 1992
|
|
1ITI
 
 | | THE HIGH RESOLUTION THREE-DIMENSIONAL SOLUTION STRUCTURE OF HUMAN INTERLEUKIN-4 DETERMINED BY MULTI-DIMENSIONAL HETERONUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | INTERLEUKIN-4 | | Authors: | Clore, G.M, Powers, B, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1993-04-12 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 32, 1993
|
|
1IVQ
 
 | |
1THC
 
 | | CRYSTAL STRUCTURE DETERMINATION AT 2.3A OF HUMAN TRANSTHYRETIN-3',5'-DIBROMO-2',4,4',6-TETRA-HYDROXYAURONE COMPLEX | | Descriptor: | 3',5'-DIBROMO-2',4,4',6'-TETRAHYDROXY AURONE, TRANSTHYRETIN | | Authors: | Ciszak, E, Cody, V, Luft, J.R. | | Deposit date: | 1992-04-20 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure determination at 2.3-A resolution of human transthyretin-3',5'-dibromo-2',4,4',6-tetrahydroxyaurone complex.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1TLA
 
 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1TDB
 
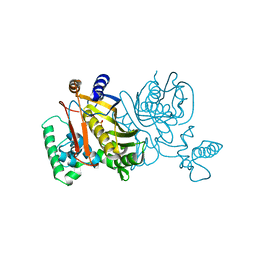 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-02-15 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
119D
 
 | |
122D
 
 | |
