4X1N
 
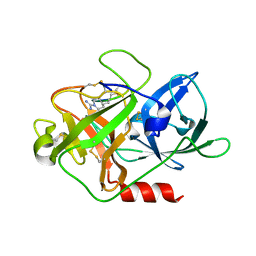 | | The crystal structure of mupain-1-16 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
8SDF
 
 | |
8OLS
 
 | |
6YCX
 
 | | Plasmodium falciparum Myosin A full-length, pre-powerstroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
1BBC
 
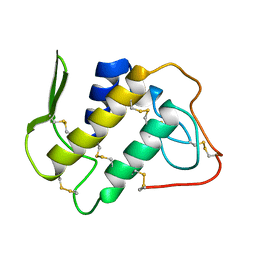 | | STRUCTURE OF RECOMBINANT HUMAN RHEUMATOID ARTHRITIC SYNOVIAL FLUID PHOSPHOLIPASE A2 AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Wery, J.-P, Schevitz, R.W, Clawson, D.K, Bobbitt, J.L, Dow, E.R, Gamboa, G, Goodsonjunior, T, Hermann, R.B, Kramer, R.M, Mcclure, D.B, Mihelich, E.D, Putnam, J.E, Sharp, J.D, Stark, D.H, Teater, C, Warrick, M.W, Jones, N.D. | | Deposit date: | 1992-05-04 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant human rheumatoid arthritic synovial fluid phospholipase A2 at 2.2 A resolution.
Nature, 352, 1991
|
|
8OM0
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Na+, Mg2+ and intronistat B | | Descriptor: | 3,4,5-trihydroxybenzoic acid, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2023-03-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8F72
 
 | | Phage P32 gp64- RNA polymerase | | Descriptor: | MAGNESIUM ION, TPR_REGION domain-containing protein | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-17 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Thermophage RNA polymerase
To Be Published
|
|
8F5M
 
 | | Crystal structure of P74 gp62 | | Descriptor: | Envelope glycoprotein gp62, MAGNESIUM ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-14 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tail-tape-fused virion and non-virion RNA polymerases of a thermophilic virus with an extremely long tail.
Nat Commun, 15, 2024
|
|
8OZM
 
 | |
4WX2
 
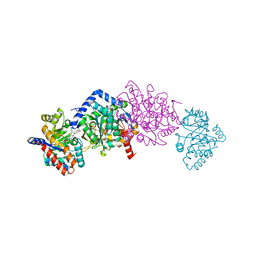 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
4WLU
 
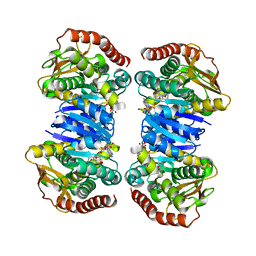 | |
4WLO
 
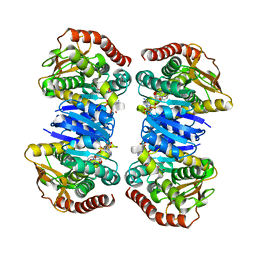 | |
8DF2
 
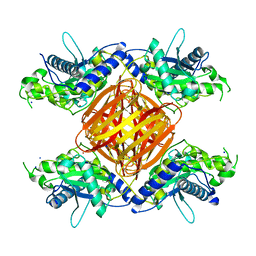 | |
6S9P
 
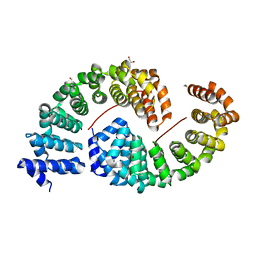 | | Designed Armadillo Repeat protein internal Lock2 fused to target peptide KRKAKITWKR | | Descriptor: | 1,2-ETHANEDIOL, internal Lock2 fused to target peptide KRKAKITWKR | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
8DEK
 
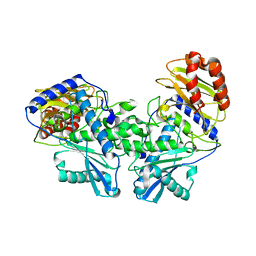 | |
8FCC
 
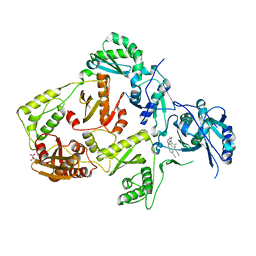 | | HIV-1 Reverse Transcriptase in complex with 5-membered bicyclic core NNRTI | | Descriptor: | 4-[(9-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-8-oxo-8,9-dihydro-7H-purin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
5IQP
 
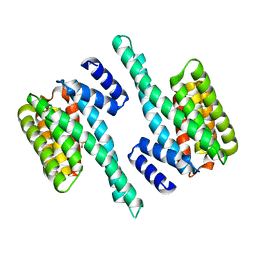 | | 14-3-3 PROTEIN TAU ISOFORM | | Descriptor: | 14-3-3 protein theta, SULFATE ION | | Authors: | Xiao, B, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways
Nature, 376, 1995
|
|
8FCD
 
 | | HIV-1 Reverse Transcriptase in complex with 6-membered bicyclic core NNRTI | | Descriptor: | 4-[(8-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-6-oxo-5,6,7,8-tetrahydropteridin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
1BE0
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING ACETIC ACID | | Descriptor: | ACETATE ION, ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
7TFB
 
 | | P. polymyxa GS(14)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
4X1Q
 
 | | The crystal structure of mupain-1 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
7AGV
 
 | | High-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETATE ION, CALCIUM ION, ... | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7TF9
 
 | | L. monocytogenes GS(14)-Q-GlnR peptide | | Descriptor: | C-tail peptide of Glutamine synthetase repressor, GLUTAMINE, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
