5H9E
 
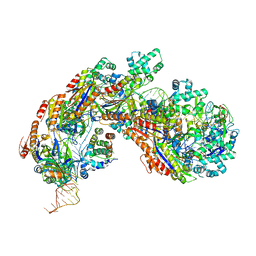 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target (32-nt spacer) at 3.20 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
8HWB
 
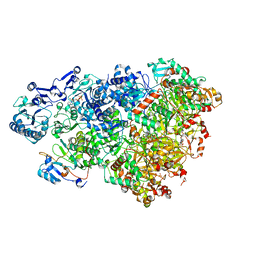 | | D5 ATP-ADP-Apo-ssDNA IS2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6S3M
 
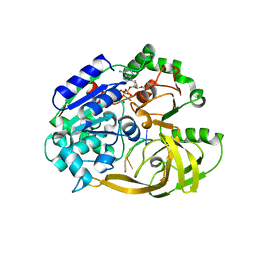 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
2B1D
 
 | |
6K08
 
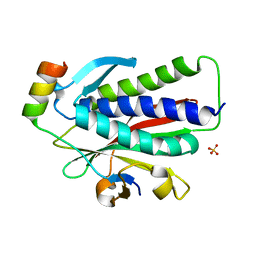 | | Crystal structure of REV7(R124A/A135D) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
4L5R
 
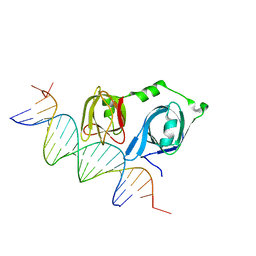 | |
4L60
 
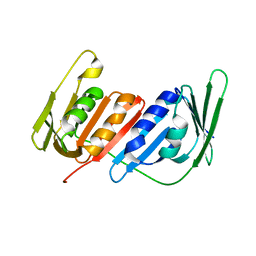 | |
4LEV
 
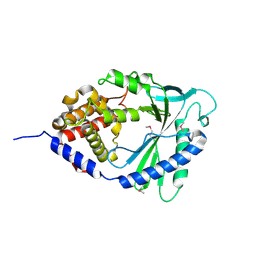 | | Structure of human cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
2EDU
 
 | | Solution structure of RSGI RUH-070, a C-terminal domain of kinesin-like protein KIF22 from human cDNA | | Descriptor: | Kinesin-like protein KIF22 | | Authors: | Abe, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-070, a C-terminal domain of kinesin-like protein KIF22 from human cDNA
To be Published
|
|
4ZVK
 
 | | Reduced quinone reductase 2 in complex with ethidium | | Descriptor: | ETHIDIUM, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | Binding of DNA-Intercalating Agents to Oxidized and Reduced Quinone Reductase 2.
Biochemistry, 54, 2015
|
|
4ZVN
 
 | | Reduced quinone reductase 2 in complex with acridine orange | | Descriptor: | ACRIDINE ORANGE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Binding of DNA-Intercalating Agents to Oxidized and Reduced Quinone Reductase 2.
Biochemistry, 54, 2015
|
|
4ZVM
 
 | | Oxidized quinone reductase 2 in complex with doxorubicin | | Descriptor: | DOXORUBICIN, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Binding of DNA-Intercalating Agents to Oxidized and Reduced Quinone Reductase 2.
Biochemistry, 54, 2015
|
|
4ZVL
 
 | | Oxidized quinone reductase 2 in complex with acridine orange | | Descriptor: | ACRIDINE ORANGE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2015-05-18 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Binding of DNA-Intercalating Agents to Oxidized and Reduced Quinone Reductase 2.
Biochemistry, 54, 2015
|
|
1KFT
 
 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
4PW7
 
 | | structure of UHRF2-SRA in complex with a 5mC-containing DNA | | Descriptor: | 5mC-containing DNA1, 5mC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
7DUV
 
 | | Structure of Sulfolobus solfataricus SegB protein | | Descriptor: | SULFATE ION, SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
4D1Q
 
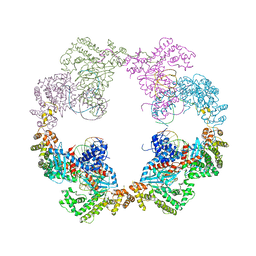 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
4LEW
 
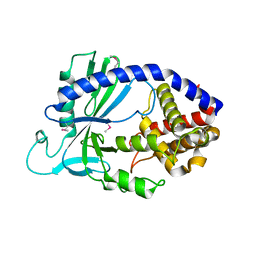 | | Structure of human cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
4L5S
 
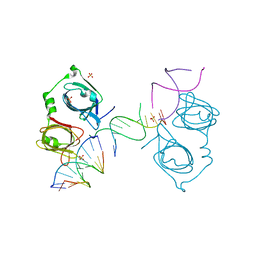 | | p202 HIN1 in complex with 12-mer dsDNA | | Descriptor: | 12-mer DNA, Interferon-activable protein 202, SULFATE ION | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
6K07
 
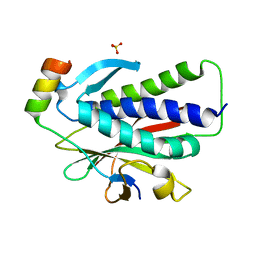 | | Crystal structure of REV7(R124A) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
4PW6
 
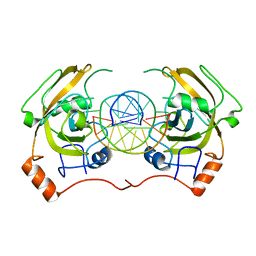 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4PW5
 
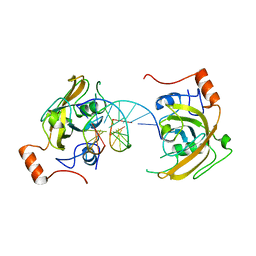 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex I | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
3S5B
 
 | |
1KQ8
 
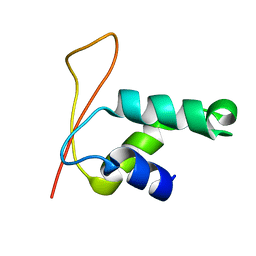 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
