7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AGA
 
 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6O9M
 
 | | Structure of the human apo TFIIH | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4S0W
 
 | | Wild type T4 lysozyme structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, E.H, Snell, M.E. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Wild type T4 lysozyme structure
To be Published
|
|
7AST
 
 | | Apo Human RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Ramsay, E.P, Abascal-Palacios, G, Daiss, J.L, King, H, Gouge, J, Pilsl, M, Beuron, F, Morris, E, Gunkel, P, Engel, C, Vannini, A. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of human RNA polymerase III.
Nat Commun, 11, 2020
|
|
7ANS
 
 | | Structure of SARS-CoV-2 Main Protease bound to Adrafinil. | | Descriptor: | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6QX3
 
 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6S6H
 
 | | Crystal structure of the DNA binding domain of the chromosome-partitioning protein ParB complexed to the centromeric parS site | | Descriptor: | Chromosome-partitioning protein ParB, DNA (5'-D(*GP*AP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*TP*C)-3'), GLYCEROL | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Tan, E.X, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2019-07-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
7AVD
 
 | | Structure of SARS-CoV-2 Main Protease bound to SEN1269 ligand | | Descriptor: | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol, 3C-like proteinase, CHLORIDE ION | | Authors: | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
7UGB
 
 | |
7AOL
 
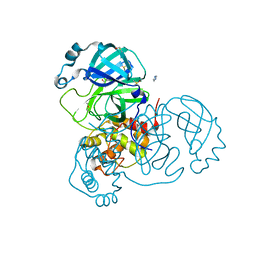 | | Structure of SARS-CoV-2 Main Protease bound to Climbazole | | Descriptor: | (1~{S})-1-(4-chloranylphenoxy)-1-imidazol-1-yl-3,3-dimethyl-butan-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1H5U
 
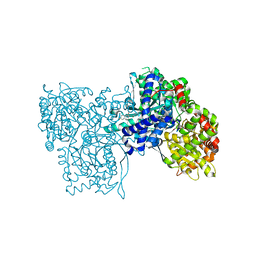 | | THE 1.76 A RESOLUTION CRYSTAL STRUCTURE OF GLYCOGEN PHOSPHORYLASE B COMPLEXED WITH GLUCOSE AND CP320626, A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Archontis, G. | | Deposit date: | 2001-05-25 | | Release date: | 2001-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The 1.76 A Resolution Crystal Structure of Glycogen Phosphorylase B Complexed with Glucose, and Cp320626, a Potential Antidiabetic Drug
Bioorg.Med.Chem., 10, 2002
|
|
4A04
 
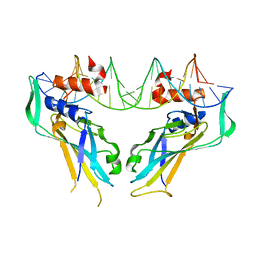 | | Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome | | Descriptor: | DNA, T-BOX TRANSCRIPTION FACTOR TBX1 | | Authors: | El Omari, K, De Mesmaeker, J, Karia, D, Ginn, H, Bhattacharya, S, Mancini, E.J. | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the DNA-Bound T-Box Domain of Human Tbx1, a Transcription Factor Associated with the Digeorge Syndrome
Proteins, 80, 2012
|
|
6R60
 
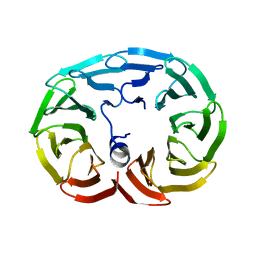 | |
1GUE
 
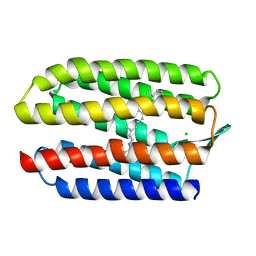 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
8U1T
 
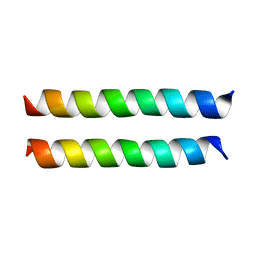 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
5VMX
 
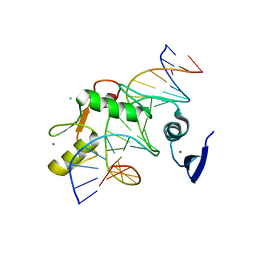 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a hemi CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*CP*GP*CP*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
8U1K
 
 | | Cryo-EM of Caulobacter crescentus Tad pilus | | Descriptor: | PilA | | Authors: | Sonani, R.R, Sanchez, J.C, Baumgardt, J.K, Wright, E.R, Egelman, E.H. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tad and toxin-coregulated pilus structures reveal unexpected diversity in bacterial type IV pili.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6RR7
 
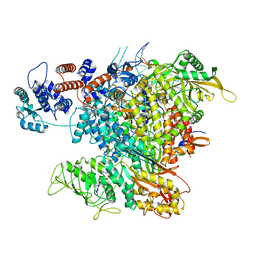 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6XKY
 
 | | Caulobacter crescentus FljK filament, straightened | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
1HB1
 
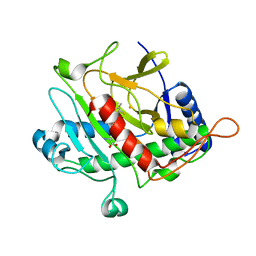 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1R)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOMETHYL)-2-OXOETHYL]-6-OXO-D-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
5NW9
 
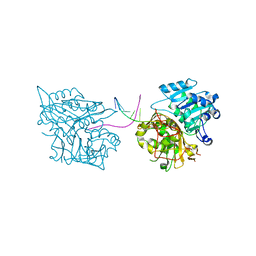 | |
6S2Z
 
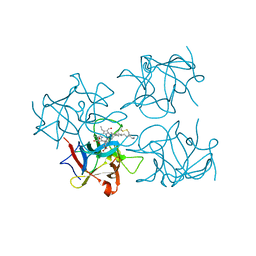 | | Water-soluble Chlorophyll Protein (WSCP) from Brassica oleracea var. Botrytis with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-Soluble Chlorophyll Protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
5NVU
 
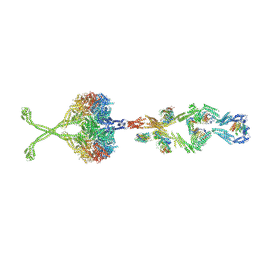 | | Full length human cytoplasmic dynein-1 in the phi-particle conformation | | Descriptor: | Dynein intermediate chain, Dynein light intermediate chain, Dynein motor domain, ... | | Authors: | Zhang, K, Foster, H.E, Carter, A.P. | | Deposit date: | 2017-05-04 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-06 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated.
Cell, 169, 2017
|
|
