6RIG
 
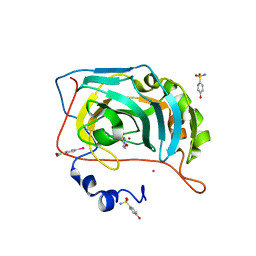 | |
6YK4
 
 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound ( S) - 1- [2'-Amino-2'-carboxyethyl]-6-methyl-5 ,7- dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 1.00A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
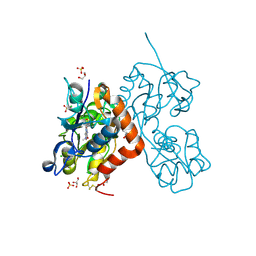
5MTU
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
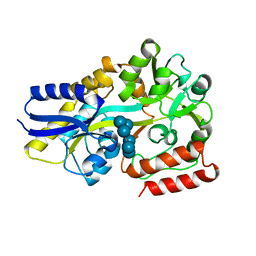
5R34
 
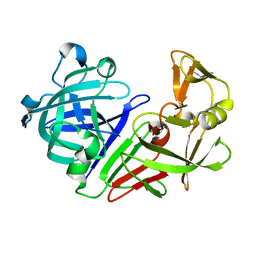 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 28, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2L
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 09, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7K4T
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5WHP
 
 | | Crystal structure of the segment, NFGTFS, from the A315T familial variant of the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | Segment of TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8TSX
 
 | |
1MN8
 
 | | Structure of Moloney Murine Leukaemia Virus Matrix Protein | | Descriptor: | Core protein p15 | | Authors: | Riffel, N, Harlos, K, Iourin, O, Rao, Z, Kingsman, A, Stuart, D, Fry, E. | | Deposit date: | 2002-09-05 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of Moloney murine leukaemia virus matrix protein and its relationship to other retroviral matrix proteins.
Structure, 10, 2002
|
|
1EXR
 
 | |
131D
 
 | | THE LOW-TEMPERATURE CRYSTAL STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA REVEALS BINDING OF A SPERMINE MOLECULE IN THE MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SODIUM ION, SPERMINE | | Authors: | Bancroft, D, Williams, L.D, Rich, A, Egli, M. | | Deposit date: | 1993-06-18 | | Release date: | 1993-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove.
Biochemistry, 33, 1994
|
|
5LXW
 
 | |
3DJK
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0255A | | Descriptor: | (5R)-1,3-dioxepan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-23 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies
J.Med.Chem., 51, 2008
|
|
7ADR
 
 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7UYD
 
 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | Descriptor: | Microtubule-associated protein tau | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
1EB6
 
 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4CNN
 
 | | High resolution structure of Salmonella typhi type I dehydroquinase | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanistic Insight Into the Reaction Catalyzed by Type I Dehydroquinase
To be Published
|
|
4UBY
 
 | |
4AXO
 
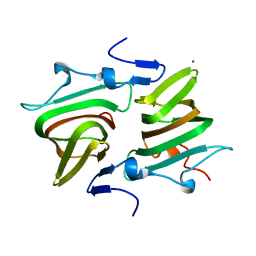 | | Structure of the Clostridium difficile EutQ protein | | Descriptor: | ETHANOLAMINE UTILIZATION PROTEIN, MAGNESIUM ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
5E1K
 
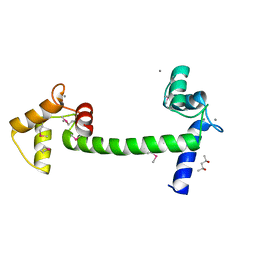 | | Selenomethionine Ca2+-Calmodulin from Paramecium tetraurelia SAD data | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2RBK
 
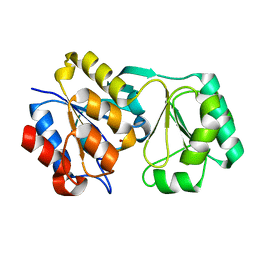 | |
4U2W
 
 | | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin | | Descriptor: | 1,2-ETHANEDIOL, Bowman-Birk trypsin inhibitor, CALCIUM ION, ... | | Authors: | Grudnik, P, Golik, P, Malicki, S, Debowski, D, Legowska, A, Rolka, K, Dubin, G. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin.
Proteins, 83, 2015
|
|
5HQ1
 
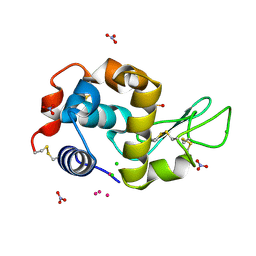 | | Comment on S. W. M. Tanley and J. R. Helliwell Structural dynamics of cisplatin binding to histidine in a protein Struct. Dyn. 1, 034701 (2014) regarding the refinement of 4mwk, 4mwm, 4mwn and 4oxe and the method we have adopted. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Helliwell, J.R. | | Deposit date: | 2016-01-21 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn, 3, 2016
|
|
6DIX
 
 | | NFVFGT segment from Human Immunoglobulin Light-Chain Variable Domain, Residues 98-103, assembled as an amyloid fibril | | Descriptor: | NFVFGT Immunoglobulin Light-Chain Variable Domain | | Authors: | Brumshtein, B, Esswein, S.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of two principal amyloid-driving segments in variable domains of Ig light chains in systemic light-chain amyloidosis.
J. Biol. Chem., 293, 2018
|
|
