2BC5
 
 | | Crystal structure of E. coli cytochrome b562 with engineered c-type heme linkages | | Descriptor: | HEME C, SULFATE ION, Soluble cytochrome b562 | | Authors: | Faraone-Mennella, J, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stability and Folding Kinetics of Structurally Characterized Cytochrome c-b(562).
Biochemistry, 45, 2006
|
|
1FQ2
 
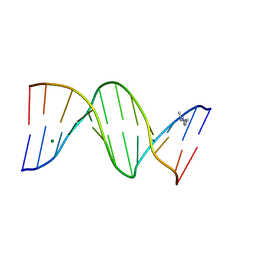 | | CRYSTAL STRUCTURE ANALYSIS OF THE POTASSIUM FORM OF B-DNA DODECAMER CGCGAATTCGCG | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Williams, L.D, Sines, C.C, McFail-Isom, L, Howerton, S.B, VanDerveer, D. | | Deposit date: | 2000-09-01 | | Release date: | 2000-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cations Mediate B-DNA Conformational Heterogeneity
J.Am.Chem.Soc., 122, 2000
|
|
2G95
 
 | |
2A83
 
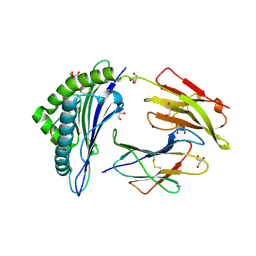 | | Crystal structure of hla-b*2705 complexed with the glucagon receptor (gr) peptide (residues 412-420) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Ruckert, C, Fiorillo, M.T, Loll, B, Moretti, R, Biesiadka, J, Saenger, W, Ziegler, A, Sorrentino, R, Uchanska-Ziegler, B. | | Deposit date: | 2005-07-07 | | Release date: | 2005-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational dimorphism of self-peptides and molecular mimicry in a disease-associated HLA-B27 subtype.
J.Biol.Chem., 281, 2006
|
|
4L4V
 
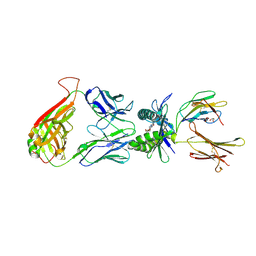 | | Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
3PQU
 
 | |
2G97
 
 | |
3VE1
 
 | |
3Q3X
 
 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | Descriptor: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | Authors: | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
4GJ0
 
 | | Crystal structure of CD23 lectin domain mutant S252A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-09 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
4GK1
 
 | | Crystal structure of CD23 lectin domain mutant D270A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
1W1T
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(His-L-Pro) at 1.9 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
4J6P
 
 | |
3BP4
 
 | | The high resolution crystal structure of HLA-B*2705 in complex with a Cathepsin A signal sequence peptide pCatA | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
1UXW
 
 | | CRYSTAL STRUCTURE OF HLA-B*2709 COMPLEXED WITH THE LATENT MEMBRANE PROTEIN 2 PEPTIDE (LMP2) OF EPSTEIN-BARR VIRUS | | Descriptor: | BETA-2-MICROGLOBULIN, GENE TERMINAL PROTEIN (MEMBRANE PROTEIN LMP-2A/LMP-2B), GLYCEROL, ... | | Authors: | Hulsmeyer, M, Kozerski, C, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-03-01 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Allele-Dependent Similarity between Viral and Self-Peptide Presentation by Hla-B27 Subtypes
J.Biol.Chem., 280, 2005
|
|
1UXS
 
 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE LATENT MEMBRANE PROTEIN 2 PEPTIDE (LMP2)OF EPSTEIN-BARR VIRUS | | Descriptor: | BETA-2-MICROGLOBULIN, GENE TERMINAL PROTEIN (MEMBRANE PROTEIN LMP-2A/LMP-2B), GLYCEROL, ... | | Authors: | Hulsmeyer, M, Kozerski, C, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-03-01 | | Release date: | 2004-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Allele-Dependent Similarity between Viral and Self-Peptide Presentation by Hla-B27 Subtypes
J.Biol.Chem., 280, 2005
|
|
1PYG
 
 | |
2G96
 
 | |
3BP7
 
 | | The high resolution crystal structure of HLA-B*2709 in complex with a Cathepsin A signal sequence peptide, pCatA | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
3BXN
 
 | | The high resolution crystal structure of HLA-B*1402 complexed with a Cathepsin A signal sequence peptide, pCatA | | Descriptor: | Cathepsin A signal sequence octapeptide, GLYCEROL, HLA-B*1402 extracellular domain, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2008-01-14 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
3VE2
 
 | |
2I72
 
 | | AmpC beta-lactamase in complex with 5-diformylaminomethyl-benzo[b]thiophen-2-boronic acid | | Descriptor: | Beta-lactamase, {5-[(DIFORMYLAMINO)METHYL]-1-BENZOTHIEN-2-YL}BORONIC ACID | | Authors: | Venturelli, A, Cancian, L, Tondi, D, Morandi, F, Cannazza, G, Segatore, B, Prati, F, Amicosante, G, Shoichet, B.K, Costi, M.P. | | Deposit date: | 2006-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimizing Cell Permeation of an Antibiotic Resistance Inhibitor for Improved Efficacy
J.Med.Chem., 50, 2007
|
|
4CP3
 
 | | The structure of BCL6 BTB (POZ) domain in complex with the ansamycin antibiotic rifabutin. | | Descriptor: | B-CELL LYMPHOMA 6 PROTEIN, RIFABUTIN | | Authors: | Evans, S.E, Fairall, L, Goult, B.T, Jamieson, A.G, Ferrigno, P.K, Ford, R, Wagner, S.D, Schwabe, J.W.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Ansamycin Antibiotic, Rifamycin Sv, Inhibits Bcl6 Transcriptional Repression and Forms a Complex with the Bcl6-Btb/Poz Domain.
Plos One, 9, 2014
|
|
7P3V
 
 | | B-Raf V600E structure bound to a new inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-[5-(2-azanylpyrimidin-4-yl)-2-[(3~{S})-morpholin-3-yl]-1,3-thiazol-4-yl]-2-fluoranyl-phenyl]-2,5-bis(fluoranyl)benzenesulfonamide | | Authors: | Schneider, M, Gelin, M, Cohen-Gonsaud, M, Labesse, G. | | Deposit date: | 2021-07-08 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based and Knowledge-Informed Design of B-Raf Inhibitors Devoid of Deleterious PXR Binding.
J.Med.Chem., 65, 2022
|
|
