3A17
 
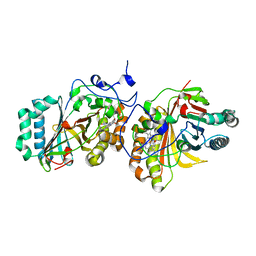 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (Co-crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
4N6R
 
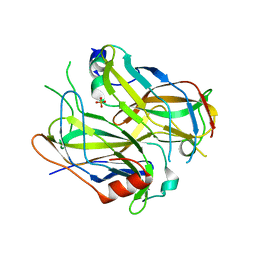 | | Crystal structure of VosA-VelB-complex | | Descriptor: | SULFATE ION, VelB, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
3A7C
 
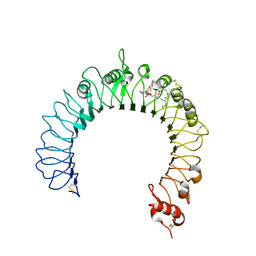 | | Crystal structure of TLR2-PE-DTPA complex | | Descriptor: | (10S,13R)-3-{2-[{2-[bis(carboxymethyl)amino]ethyl}(carboxymethyl)amino]ethyl}-10-hydroxy-5,16-dioxo-13-(tetradecanoyloxy)-9,11,15-trioxa-3,6-diaza-10-phosphanonacosan-1-oic acid 10-oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kang, J.Y, Jin, M.S, Lee, J.-O. | | Deposit date: | 2009-09-20 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer
Immunity, 31, 2009
|
|
3A4I
 
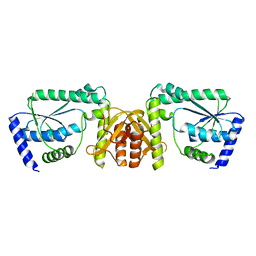 | | Crystal structure of GMP synthetase PH1347 from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B | | Authors: | Maruoka, S, Horita, S, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the ATPPase subunit and its substrate-dependent association with the GATase Subunit: a novel regulatory mechanism for a two-subunit-type GMP synthetase from Pyrococcus horikoshii OT3.
J.Mol.Biol., 395, 2010
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3AAE
 
 | | Crystal structure of Actin capping protein in complex with CARMIL fragment | | Descriptor: | 32mer peptide from Leucine-rich repeat-containing protein 16A, F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2 | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-16 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Two distinct mechanisms for the regulation of actin capping protein-competitive or allosteric inhibitions
To be Published
|
|
3AA0
 
 | | Crystal structure of Actin Capping Protein in complex with the Cp-binding motif derived from CARMIL | | Descriptor: | 21mer peptide from Leucine-rich repeat-containing protein 16A, CARBONATE ION, F-actin-capping protein subunit alpha-1, ... | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
3AE8
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with N-(3-Isopropoxy-phenyl)-2-trifluoromethylbenzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with N-(3-Isopropoxy-phenyl)-2-trifluoromethylbenzamide
To be Published
|
|
3AG1
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 280 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4N8D
 
 | | DPP4 complexed with syn-7aa | | Descriptor: | 1-(cis-1-phenyl-4-{[(2E)-3-phenylprop-2-en-1-yl]oxy}cyclohexyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of C-(1-aryl-cyclohexyl)-methylamines as selective, orally available inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3A3V
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
3A0X
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 1: ammomium phosphate, monoclinic) | | Descriptor: | Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A15
 
 | | Crystal Structure of Substrate-Free Form of Aldoxime Dehydratase (OxdRE) | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
4N9E
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3A5L
 
 | | Crystal Structure of a Dictyostelium P109A Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
4NKP
 
 | |
4Q07
 
 | | Crystal structure of chimeric carbonic anhydrase IX with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4PZI
 
 | | Zinc finger region of MLL2 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3'), Histone-lysine N-methyltransferase 2B, UNKNOWN ATOM OR ION, ... | | Authors: | Chao, X, Tempel, W, Liu, K, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4Q06
 
 | | Crystal structure of chimeric carbonic anhydrase IX with inhibitor | | Descriptor: | 2-(cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q5K
 
 | | Crystal structure of a N-acetylmuramoyl-L-alanine amidase (BACUNI_02947) from Bacteroides uniformis ATCC 8492 at 1.30 A resolution | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, SODIUM ION, Uncharacterized protein | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-guided functional characterization of DUF1460 reveals a highly specific NlpC/P60 amidase family.
Structure, 22, 2014
|
|
4Q9K
 
 | | P-glycoprotein cocrystallised with QZ-Leu | | Descriptor: | (30F)L(30F)L(30F)L Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q53
 
 | | Crystal structure of a DUF4783 family protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CHLORIDE ION, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-16 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of a hypothetical protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution
To be published
|
|
4Q98
 
 | |
4Q6D
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-[(Z)-azepan-1-yldiazenyl]benzenesulfonamide | | Descriptor: | 4-[(E)-azepan-1-yldiazenyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 4-Amino-substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Molecules, 19, 2014
|
|
4PW1
 
 | |
