1KDH
 
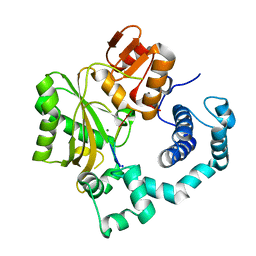 | | Binary Complex of Murine Terminal Deoxynucleotidyl Transferase with a Primer Single Stranded DNA | | Descriptor: | 5'-D(P*(BRU)P*(BRU)P*(BRU)P*(BRU))-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
8ATE
 
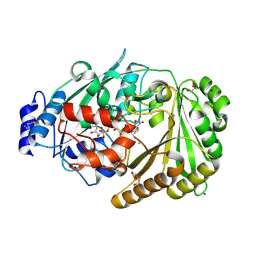 | | Galacturonic acid oxidase from Citrus sinensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boverio, A, Savino, S, Fraaije, M.W, Loncar, N. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and Structural Characterization of a Uronic Acid Oxidase from Citrus sinensis
Chemcatchem, 2023
|
|
8U4F
 
 | |
8U4A
 
 | |
8U49
 
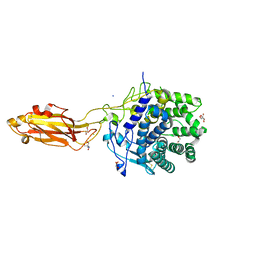 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8U5R
 
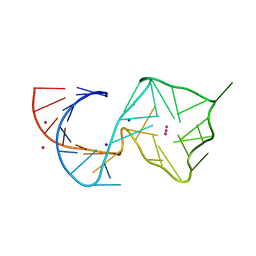 | |
8AGQ
 
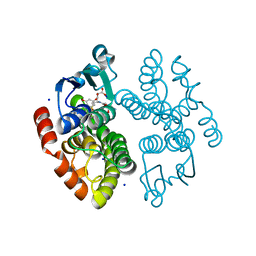 | | Crystal structure of anthocyanin-related GSTF8 from Populus trichocarpa in complex with (-)-catechin and glutathione | | Descriptor: | (2~{S},3~{R})-2-[3,4-bis(oxidanyl)phenyl]-3,4-dihydro-2~{H}-chromene-3,5,7-triol, GLUTATHIONE, Glutathione transferase, ... | | Authors: | Eichenberger, M, Hueppi, S, Schwander, T, Mittl, P, Buller, M.R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.093 Å) | | Cite: | The catalytic role of glutathione transferases in heterologous anthocyanin biosynthesis.
Nat Catal, 6, 2023
|
|
8U5T
 
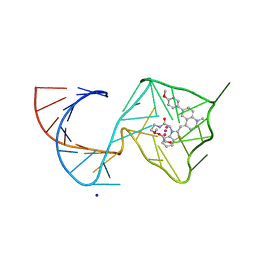 | | Structure of Mango II variant aptamer bound to T01-6A-B | | Descriptor: | 3-{2,16-dioxo-20-[(3aS,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-6A-B
To Be Published
|
|
7RAX
 
 | |
7REC
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to 5-HDC | | Descriptor: | 5-hydroxydiclofenac, Calcium/calmodulin-dependent protein kinase type II subunit alpha, SODIUM ION | | Authors: | McSpadden, E.D, Chi, C.C, Gee, C.L, Kuriyan, J. | | Deposit date: | 2021-07-12 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GHB analogs confer neuroprotection through specific interaction with the CaMKII alpha hub domain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8TW1
 
 | | Crystal structure of Lys2972, a phage endolysin targeting Streptococcus thermophilus | | Descriptor: | Endolysin Lys2972, GLYCEROL, SODIUM ION | | Authors: | Zhu, X, Moineau, S, Shi, R. | | Deposit date: | 2023-08-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Fermentation Practices Select for Thermostable Endolysins in Phages.
Mol.Biol.Evol., 41, 2024
|
|
8AGP
 
 | | Halogenated product of limonene epoxide turnover by epoxide hydrolase from metagenomic source ch65 | | Descriptor: | (1~{S},2~{S},4~{R})-2-chloranyl-1-methyl-4-prop-1-en-2-yl-cyclohexan-1-ol, 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8TZ2
 
 | |
8AJ0
 
 | | Mpro of SARS COV-2 in complex with the RK-90 inhibitor | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor RK-90
To Be Published
|
|
8U5Z
 
 | | Structure of Mango II variant aptamer bound to T01-7M-B | | Descriptor: | 2-[(E)-(1,7-dimethylquinolin-4(1H)-ylidene)methyl]-3-{2,16-dioxo-20-[(3aR,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-7M-B
To Be Published
|
|
8QWR
 
 | | Crystal structure of CotB2 variant V80L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclooctat-9-en-7-ol synthase, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CotB2 variant V80L
To Be Published
|
|
8TZ1
 
 | | Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, ... | | Authors: | Wright, N.J, Lee, S.-Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Antiviral drug recognition and elevator-type transport motions of CNT3.
Nat.Chem.Biol., 20, 2024
|
|
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
8QD6
 
 | | AntI Ser245DHA (PMSF) | | Descriptor: | ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, SODIUM ION, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QHK
 
 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to reduced 3-acetylpyridine adenine dinucleotide | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, ... | | Authors: | Wohlwend, D, Friedrich, T, Bucka, S. | | Deposit date: | 2023-09-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
8QG1
 
 | |
8TZ5
 
 | |
8TZ6
 
 | |
8QH4
 
 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to oxidized 3-acetylpyridine adenine dinucleotide | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
8QCU
 
 | | Lysozyme covalently bound to fac-[Re(CO)3-imidazole] complex, incubated for 38 weeks. | | Descriptor: | BROMIDE ION, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Jacobs, F.J.F, Brink, A, Helliwell, J.R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Time-series analysis of rhenium(I) organometallic covalent binding to a model protein for drug development.
Iucrj, 11, 2024
|
|
