4LHI
 
 | |
4LGQ
 
 | |
4LJX
 
 | |
4JD2
 
 | |
4JHY
 
 | |
4JQT
 
 | |
4J8P
 
 | |
4L8J
 
 | |
4LBA
 
 | |
4LDS
 
 | |
4LII
 
 | |
4LIR
 
 | |
4LG9
 
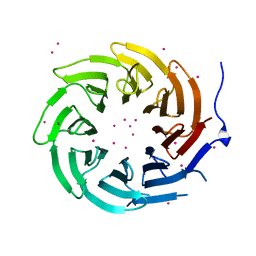 | | Crystal structure of TBL1XR1 WD40 repeats | | Descriptor: | F-box-like/WD repeat-containing protein TBL1XR1, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, He, H, Wu, X, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of TBL1XR1 WD40 repeats
TO BE PUBLISHED
|
|
4J8Q
 
 | |
4JH0
 
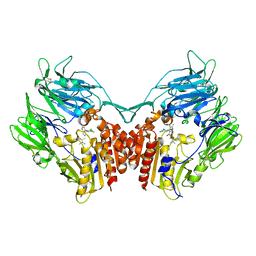 | | Crystal structure of dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) (DPP-IV-WT) complex with bms-767778 AKA 2-(3-(aminomethyl)-4-(2,4- dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6h-pyrrolo[3,4- b]pyridin-6-yl)-n,n-dimethylacetamide | | Descriptor: | 2-[3-(aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-N,N-dimethylacetamide, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
4JX0
 
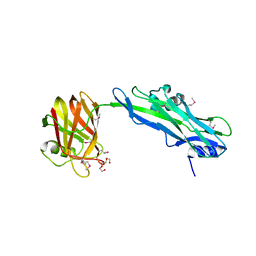 | |
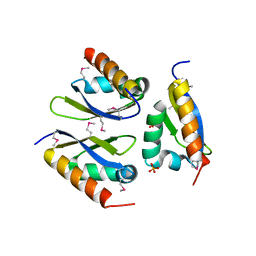
4LIJ
 
 | |
4LG3
 
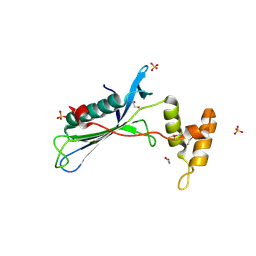 | |
4LG8
 
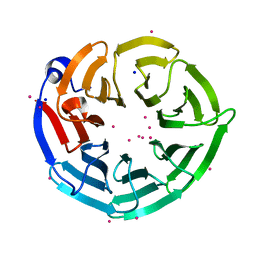 | | Crystal structure of PRPF19 WD40 repeats | | Descriptor: | Pre-mRNA-processing factor 19, SODIUM ION, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, He, H, Dobrovetsky, E, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the WD40 domain of human PRPF19.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
4LGC
 
 | |
4LEH
 
 | |
4LG7
 
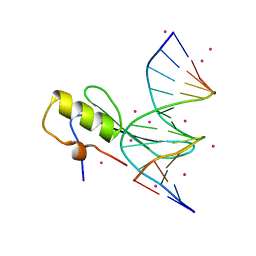 | | Crystal structure MBD4 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 4, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure MBD4 MBD domain in complex with methylated CpG DNA
To be Published
|
|
4LGJ
 
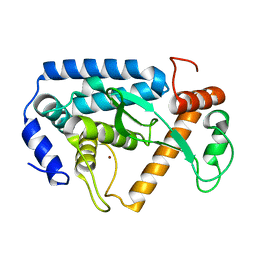 | | Crystal structure and mechanism of a type III secretion protease | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4K70
 
 | |
4K24
 
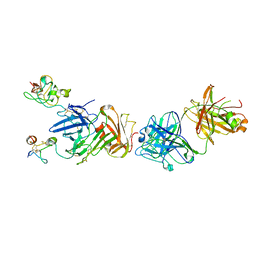 | | Structure of anti-uPAR Fab ATN-658 in complex with uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, ... | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
