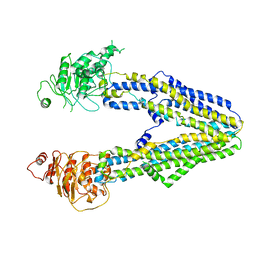
8JVH
 
 | |
5NVP
 
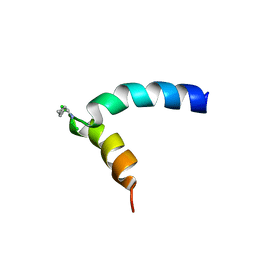 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
8UP3
 
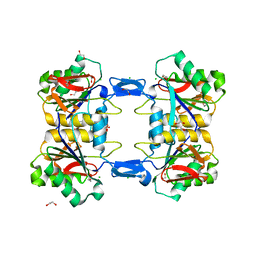 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
7NB7
 
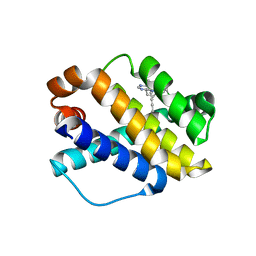 | | Structure of Mcl-1 complex with compound 6b | | Descriptor: | (2~{R})-2-[[7-but-2-ynyl-5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
8ZUV
 
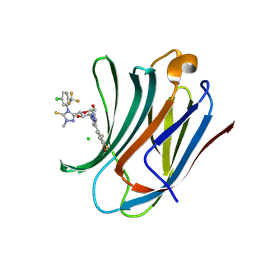 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 5-[(2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-bromanyl-2,3-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]-4-[5-chloranyl-2-(trifluoromethyl)phenyl]-2-methyl-1,2,4-triazole-3-thione, CHLORIDE ION, Galectin-3 | | Authors: | Amit, K, Swetha, R, Ghosh, K. | | Deposit date: | 2024-06-10 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atropisomerism Observed in Galactose-Based Monosaccharide Inhibitors of Galectin-3 Comprising 2-Methyl-4-phenyl-2,4-dihydro-3 H -1,2,4-triazole-3-thione.
J.Med.Chem., 67, 2024
|
|
7B4V
 
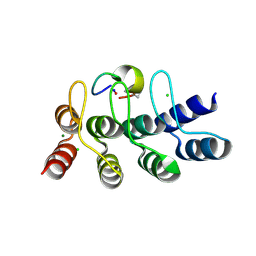 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.2, CHLORIDE ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
8WWU
 
 | | 1-naphthylamine GS in complex with AMP PNP | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a broad-substrate-spectrum enzyme catalyzing 1-naphthylamine glutamylation.
Elife, 13, 2024
|
|
9YH8
 
 | | M17T Human DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Parkinson disease protein 7 | | Authors: | Lin, J, Kovalevsky, A, Walker, A.R, Wilson, M.A. | | Deposit date: | 2025-09-30 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Environmental contributions to proton sharing in protein low barrier hydrogen bonds
To Be Published
|
|
6RG3
 
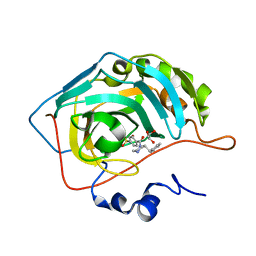 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(2~{R})-2-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
9E70
 
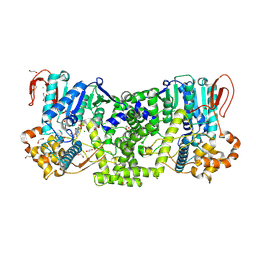 | | Human XRN1 with Adenosine-3',5'-Bisphosphate (pAp) Bound | | Descriptor: | 1,2-ETHANEDIOL, 5'-3' exoribonuclease 1, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Lockbaum, G.J, Lynes, M.M, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2024-10-31 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of exoribonuclease XRN1 as a cancer target and identification of adenosine-3',5'-bisphosphate as a potent enzyme inhibitor.
Commun Biol, 8, 2025
|
|
8GAG
 
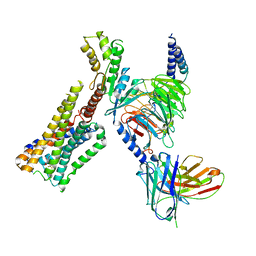 | | Cannabinoid receptor 1-Gi complex with novel ligand | | Descriptor: | Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tummino, T.A, Iliopoulos-Tsoutsouvas, C, Braz, J.M, O'Brien, E.S, Krishna Kumar, K, Makriyannis, M, Basbaum, A.I, Shoichet, B.K. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cannabinoid receptor 1-Gi complex with novel ligand
To Be Published
|
|
9JGB
 
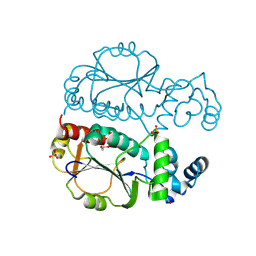 | | Crystal structure of Nep1 from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribosomal RNA small subunit methyltransferase Nep1, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
6XXT
 
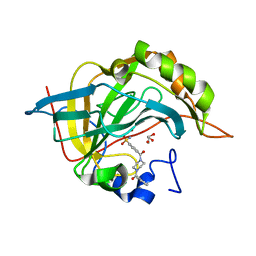 | |
6YKO
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 11a | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
7SC0
 
 | |
132L
 
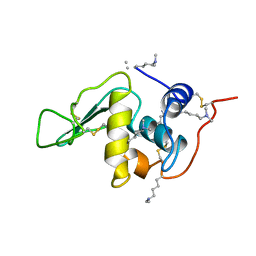 | |
8S4F
 
 | | Human carbonic anhydrase I covalently bound to AV21-08 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(cyclooctylamino)-4-ethylsulfonyl-2,5,6-tris(fluoranyl)benzenesulfonamide, ... | | Authors: | Manakova, E.N, Grazulis, S, Paketuryte-Latve, V, Vaskevicius, A, Smirnov, A. | | Deposit date: | 2024-02-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Targeted anticancer pre-vinylsulfone covalent inhibitors of carbonic anhydrase IX.
Elife, 13, 2024
|
|
8IGG
 
 | |
7NTO
 
 | | The structure of RRM domain of human TRMT2A at 1.23 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Witzenberger, M, Janowski, R, Davydova, E, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
8X41
 
 | | Crystal structure of DIMT1 in complex with 5'-methylthioadenosine and adenosine from Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ACETATE ION, ... | | Authors: | Saha, S, Mandal, S.K, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
8X3W
 
 | | Crystal structure of DIMT1 from the thermophilic archaeon, Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Sayan, S, Mandal, S.K, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
9F18
 
 | |
5ZEF
 
 | | Crystal structure of Entamoeba histolytica Arginase in complex with L- Norvaline at 2.01 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
8JTZ
 
 | | hOCT1 in complex with spironolactone in outward facing partially occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
5LJJ
 
 | | Crystal structure of human Mps1 (TTK) in complex with Reversine | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-07-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of reversine selectivity in inhibiting Mps1 more potently than aurora B kinase.
Proteins, 84, 2016
|
|
