7XVZ
 
 | |
7YE8
 
 | | Crystal structure of SARS-CoV-2 refolded dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
4AEA
 
 | | Dimeric alpha-cobratoxin X-ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, LONG NEUROTOXIN 1 | | Authors: | Rucktooa, P, Osipov, A.V, Kasheverov, I.E, Filkin, S.Y, Starkov, V.G, Andreeva, T.V, Bertrand, D, Utkin, Y.N, Tsetlin, V.I, Sixma, T.K. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dimeric Alpha-Cobratoxin X-Ray Structure: Localization of Intermolecular Disulfides and Possible Mode of Binding to Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 287, 2012
|
|
7XSW
 
 | | Structure of SARS-CoV-2 antibody S309 with GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Lambda Chain, ... | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-05-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7XWT
 
 | |
7XWV
 
 | |
7XWC
 
 | |
7YJP
 
 | | Crystal structure of MCR-1 treated by AuCl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Gold drugs as colistin adjuvants in the fight against MCR-1 producing bacteria.
J.Biol.Inorg.Chem., 28, 2023
|
|
7YJS
 
 | |
7YJR
 
 | |
7YJQ
 
 | | Crystal structure of MCR-1-S treated by auranofin | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Sun, H, Wang, M. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gold drugs as colistin adjuvants in the fight against MCR-1 producing bacteria.
J.Biol.Inorg.Chem., 28, 2023
|
|
7YJT
 
 | |
7Y9P
 
 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
7WIK
 
 | |
7W2A
 
 | | gliadinase Bga1903 | | Descriptor: | CALCIUM ION, Peptidase | | Authors: | Meng, M. | | Deposit date: | 2021-11-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Burkholderia peptidase and modification of the substrate-binding site for enhanced hydrolytic activity toward gluten-derived pro-immunogenic peptides.
Int.J.Biol.Macromol., 222, 2022
|
|
7WBB
 
 | | Cryo-EM structure of substrate engaged Drg1 hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AFG2 isoform 1, substrate | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
2WJU
 
 | |
2WJI
 
 | | Structure and function of the FeoB G-domain from Methanococcus jannaschii | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B HOMOLOG, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Koester, S, Wehner, M, Herrmann, C, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2009-05-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure and Function of the Feob G-Domain from Methanococcus Jannaschii
J.Mol.Biol., 392, 2009
|
|
2WQN
 
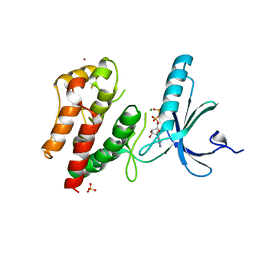 | | Structure of ADP-bound human Nek7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Richards, M.W, Bayliss, R. | | Deposit date: | 2009-08-24 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Autoinhibitory Tyrosine Motif in the Cell-Cycle- Regulated Nek7 Kinase is Released Through Binding of Nek9.
Mol.Cell, 36, 2009
|
|
7YUL
 
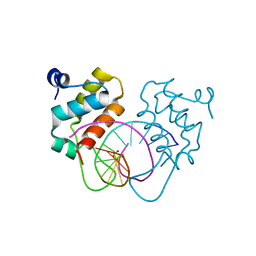 | | Crystal structure of human BEND6 BEN domain in complex with DNA | | Descriptor: | BEN domain-containing protein 6, DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCOLIC ACID | | Authors: | Liu, K, Xiao, Y.Q, Zhang, J, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
3F9L
 
 | |
7YUN
 
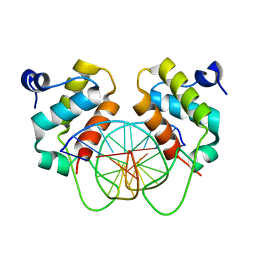 | | Crystal structure of human BEND6 BEN domain in complex with methylated DNA | | Descriptor: | BEN domain-containing protein 6, DNA (5'-D(*CP*TP*CP*TP*CP*GP*(5CM)P*GP*AP*GP*AP*G)-3') | | Authors: | Liu, K, Xiao, Y.Q, Zhang, J, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YX0
 
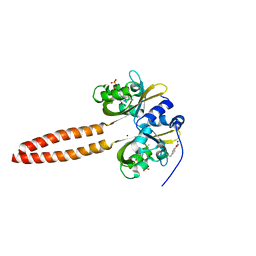 | | Crystal structure of the full-length short LOV protein SBW25-LOV from Pseudomonas fluorescens (light state) | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Flavin mononucleotide (semi-quinone intermediate), ... | | Authors: | Arinkin, V, Batra-Safferling, R, Granzin, J. | | Deposit date: | 2022-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved Signal Transduction Mechanisms and Dark Recovery Kinetic Tuning in the Pseudomonadaceae Short Light, Oxygen, Voltage (LOV) Protein Family.
J.Mol.Biol., 2024
|
|
3FCZ
 
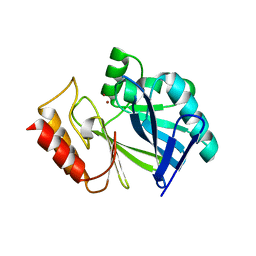 | | Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility | | Descriptor: | Beta-lactamase 2, ZINC ION | | Authors: | Tomatis, P, Fabiane, S, Simona, F, Carloni, P, Sutton, B, Vila, A. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VCV
 
 | |
