4DJO
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP56 | | Descriptor: | 2-[(dichloroacetyl)amino]ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, GLYCEROL, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
4DJQ
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP86 | | Descriptor: | 2-(2-oxoimidazolidin-1-yl)ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, PHOSPHATE ION, Pol polyprotein | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
4DJV
 
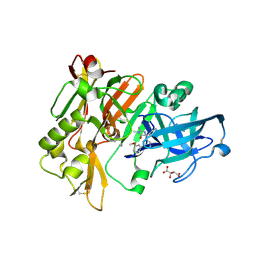 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DK8
 
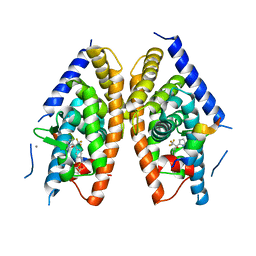 | | Crystal structure of LXR ligand binding domain in complex with partial agonist 5 | | Descriptor: | ACETATE ION, CALCIUM ION, N-methyl-N-(4-{(1S)-2,2,2-trifluoro-1-hydroxy-1-[1-(2-methoxyethyl)-1H-pyrrol-2-yl]ethyl}phenyl)benzenesulfonamide, ... | | Authors: | Piper, D.E, Kopecky, D.J, Xu, H. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of a new binding mode for a series of liver X receptor agonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DNR
 
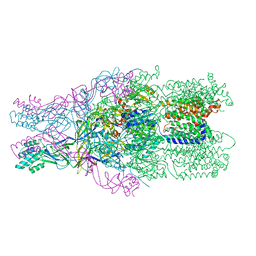 | |
2IFX
 
 | |
4DLS
 
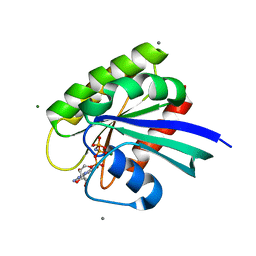 | | H-Ras Set 1 CaCl2 'Mixed' | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
3OW9
 
 | |
2IIC
 
 | |
3P69
 
 | |
3P6L
 
 | |
4DRM
 
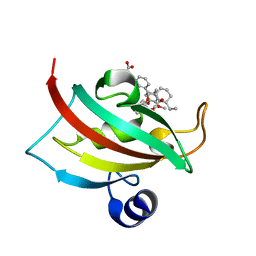 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
3OXX
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, 1,2-ETHANEDIOL, ... | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3P9K
 
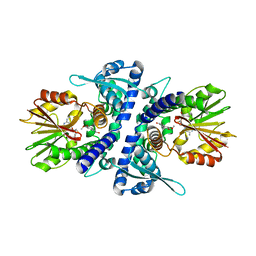 | |
4DS4
 
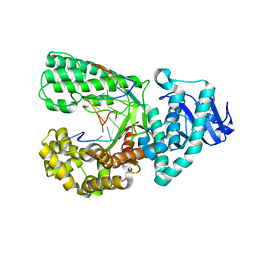 | | Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and rCTP in presence of Mn2+ | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*GP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DOC))-3'), DNA polymerase, ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
4DR5
 
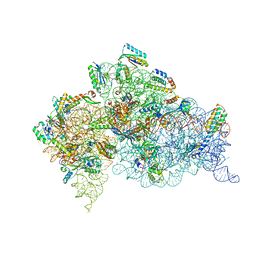 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered cognate transfer RNA anticodon stem-loop and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
3OMG
 
 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | Descriptor: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ONM
 
 | | Effector binding Domain of LysR-Type transcription factor RovM from Y. pseudotuberculosis | | Descriptor: | Transcriptional regulator LrhA | | Authors: | Quade, N, Diekmann, M, Haffke, M, Heroven, A.K, Dersch, P, Heinz, D.W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the effector-binding domain of the LysR-type transcription factor RovM from Yersinia pseudotuberculosis.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OKK
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
4DWB
 
 | | Crystal structure of Trypanosoma cruzi farnesyl diphosphate synthase in complex with [2-(n-pentylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ACETATE ION, Farnesyl pyrophosphate synthase, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
4DES
 
 | |
3MV9
 
 | | Crystal Structure of the TK3-Gln55Ala TCR in complex with HLA-B*3501/HPVG | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chen, Z, Miles, J.J, Liu, Y.C, Bell, M.J, Sullivan, L.C, Kjer-Nielsen, L, Brennan, R.M, Burrows, J.M, Neller, M.A, Khanna, R, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allelic polymorphism in the T cell receptor and its impact on immune responses
J.Exp.Med., 207, 2010
|
|
4DE2
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 12 | | Descriptor: | 3-[(dimethylamino)methyl]-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
2ICX
 
 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UTP | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE 5'-TRIPHOSPHATE | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
2ICT
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
