4M3T
 
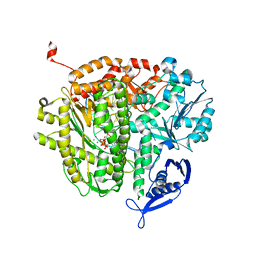 | |
4LPH
 
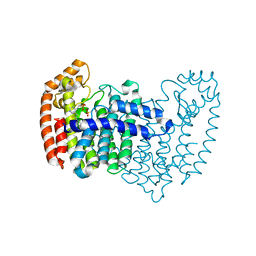 | | Crystal structure of human FPPS in complex with CL03093 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl)phosphonic acid, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
4LR6
 
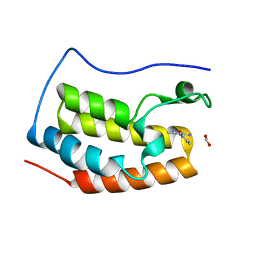 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4M3Y
 
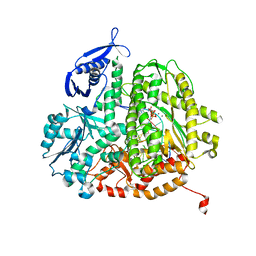 | |
4MPU
 
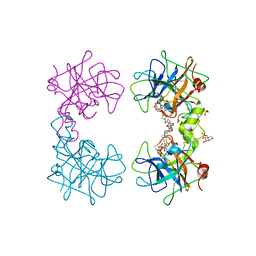 | | Human beta-tryptase co-crystal structure with (6S,8R)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-8-hydroxy-6-(1-hydroxycyclobutyl)-5,7-dioxaspiro[3.4]octane-6,8-dicarboxamide | | Descriptor: | (6S,8R)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-8-hydroxy-6-(1-hydroxycyclobutyl)-5,7-dioxaspiro[3.4]octane-6,8-dicarboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R.K. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Target-Directed Self-Assembly of Homodimeric Drugs Against beta-Tryptase.
Acs Med.Chem.Lett., 9, 2018
|
|
5Q0V
 
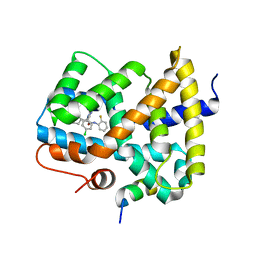 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-(2-fluorophenyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4L2X
 
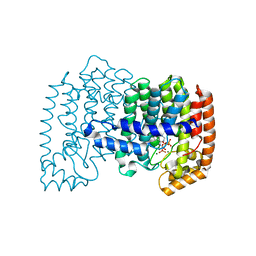 | | Crystal structure of human FPPS in complex with magnesium, CL02134, and inorganic pyrophosphate | | Descriptor: | ({[6-(4-cyclopropylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methanediyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Park, J, Leung, C.-Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Thienopyrimidine Bisphosphonate (ThPBP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase: Optimization and Characterization of the Mode of Inhibition.
J.Med.Chem., 56, 2013
|
|
5Q0P
 
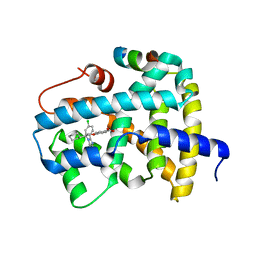 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q16
 
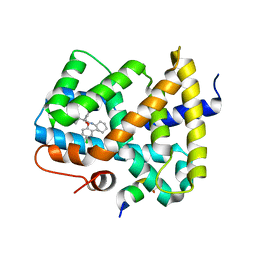 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[(4-chloro-2-methylphenoxy)methyl]-6-fluoro-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4LDW
 
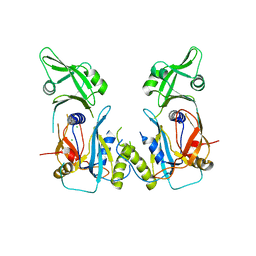 | |
4L73
 
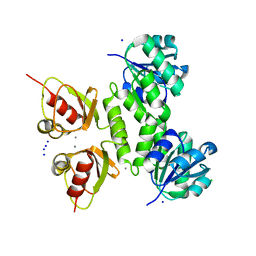 | | Ca2+-bound MthK RCK domain at 2.5 Angstrom | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calcium-gated potassium channel MthK, ... | | Authors: | Smith, F.J, Rothberg, B.S. | | Deposit date: | 2013-06-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of allosteric interactions among Ca(2+)-binding sites in a K(+) channel RCK domain.
Nat Commun, 4, 2013
|
|
4L75
 
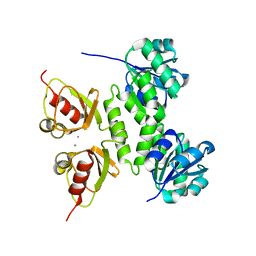 | |
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Z
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl (5S)-3-(3,4-difluorobenzene-1-carbonyl)-1,1-dimethyl-1,2,3,4,5,6-hexahydroazepino[4,5-b]indole-5-carboxylate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4LF7
 
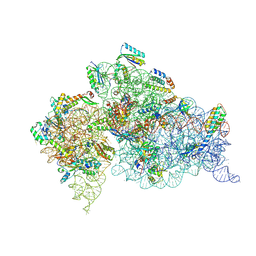 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, PAROMOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.1484 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
To be Published, 2013
|
|
4LFA
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, HYGROMYCIN B, MAGNESIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.6461 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
4LEP
 
 | | Structural insights into substrate recognition in proton dependent oligopeptide transporters | | Descriptor: | N-[(1R)-1-phosphonoethyl]-L-alaninamide, Proton:oligopeptide symporter POT family, ZINC ION | | Authors: | Guettou, F, Quistgaard, E.M, Tresaugues, L, Moberg, P, Jegerschold, C, Zhu, L, Jong, A.J, Nordlund, P, Low, C. | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into substrate recognition in proton-dependent oligopeptide transporters.
Embo Rep., 14, 2013
|
|
4LF5
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, HYGROMYCIN B, MAGNESIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.753 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
To be Published, 2013
|
|
4LFB
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, NEOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
4LFV
 
 | | Crystal structure of human FPPS in complex with YS0470 and two molecules of inorganic phosphate | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human farnesyl pyrophosphate synthase in complex with an aminopyridine bisphosphonate and two molecules of inorganic phosphate.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4LFC
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: |
to be published, 2013
|
|
4LF9
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDROXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, MAGNESIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.2811 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
4LPG
 
 | | Crystal structure of human FPPS in complex with CL01131 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methanediyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
4LQY
 
 | | Crystal Structure of Human ENPP4 with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Albright, R.A, Braddock, D.T. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis of purinergic signal metabolism by ectonucleotide pyrophosphatase/phosphodiesterases 4 and 1 and implications in stroke.
J.Biol.Chem., 289, 2014
|
|
