6T0U
 
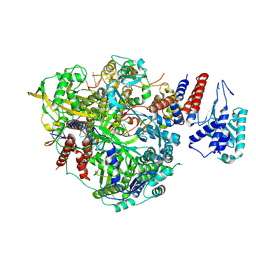 | | Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with intact oligo(U) sequence | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6T0W
 
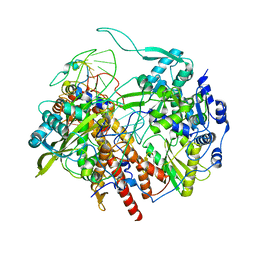 | | Human Influenza B polymerase recycling complex | | Descriptor: | Polymerase PB2, Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Karuppasamy, M, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
5EPI
 
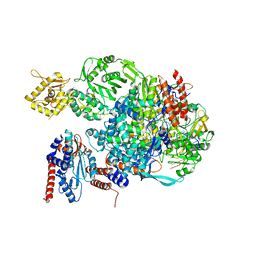 | |
2E9Z
 
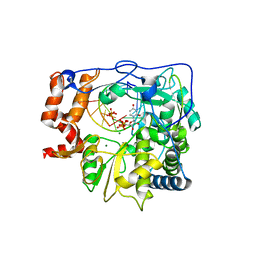 | | Foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA, ATP and UTP | | Descriptor: | 5'-R(*CP*AP*UP*GP*GP*GP*CP*CP*C)-3', 5'-R(*GP*GP*GP*CP*CP*CP*A)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-29 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7COA
 
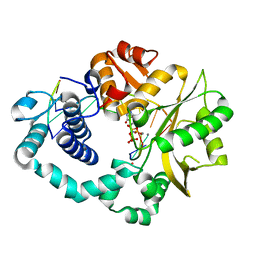 | | Ternary complex of DNA polymerase Mu with 1-nt gapped DNA (T:dGMPNPP) and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
3KYH
 
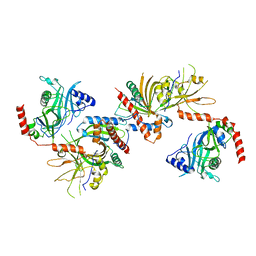 | | Saccharomyces cerevisiae Cet1-Ceg1 capping apparatus | | Descriptor: | mRNA-capping enzyme subunit alpha, mRNA-capping enzyme subunit beta | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Structure, 18, 2010
|
|
4M6T
 
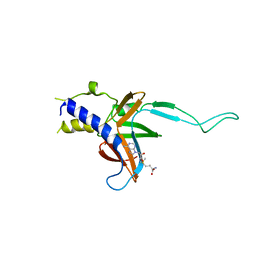 | | Structure of human Paf1 and Leo1 complex | | Descriptor: | RNA polymerase II-associated factor 1 homolog, Linker, RNA polymerase-associated protein LEO1, ... | | Authors: | Shen, Y, Qin, X. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural insights into Paf1 complex assembly and histone binding
Nucleic Acids Res., 41, 2013
|
|
2X1A
 
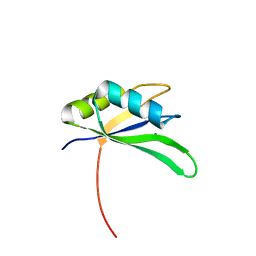 | | Structure of Rna15 RRM with RNA bound (G) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MAGNESIUM ION, MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1F
 
 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2HZS
 
 | | Structure of the Mediator head submodule Med8C/18/20 | | Descriptor: | RNA polymerase II mediator complex subunit 18, RNA polymerase II mediator complex subunit 20, RNA polymerase II mediator complex subunit 8 | | Authors: | Lariviere, L, Geiger, S, Hoeppner, S, Rother, S, Straesser, K, Cramer, P. | | Deposit date: | 2006-08-09 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YKE
 
 | | Structure of the mediator MED7/MED21 subcomplex | | Descriptor: | RNA polymerase II holoenzyme component SRB7, RNA polymerase II mediator complex protein MED7 | | Authors: | Baumli, S, Hoeppner, S, Cramer, P. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer.
J.Biol.Chem., 280, 2005
|
|
1YKH
 
 | |
6ZBK
 
 | | Crystal structure of the human complex between RPAP3 and TRBP | | Descriptor: | RISC-loading complex subunit TARBP2, RNA polymerase II-associated protein 3 | | Authors: | Charron, C, Abel, Y, Charpentier, B, Rederstorff, M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The interaction between RPAP3 and TRBP reveals a possible involvement of the HSP90/R2TP chaperone complex in the regulation of miRNA activity.
Nucleic Acids Res., 50, 2022
|
|
2HZM
 
 | | Structure of the Mediator head subcomplex Med18/20 | | Descriptor: | PHOSPHATE ION, RNA polymerase II mediator complex subunit 18, RNA polymerase II mediator complex subunit 20 | | Authors: | Lariviere, L, Geiger, S, Hoeppner, S, Rother, S, Straesser, K, Cramer, P. | | Deposit date: | 2006-08-09 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ILZ
 
 | |
6TU5
 
 | | Influenza A/H7N9 polymerase core (apo) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Cusack, S, Pflug, A. | | Deposit date: | 2020-01-02 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6T2C
 
 | | Bat Influenza A polymerase recycling complex | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-08 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6EVK
 
 | |
7UND
 
 | | Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome complex (stalled at +38) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Filipovski, M, Vos, S.M, Farnung, L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosome retention during transcription elongation.
Science, 376, 2022
|
|
7UNC
 
 | | Pol II-DSIF-SPT6-PAF1c-TFIIS complex with rewrapped nucleosome | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Filipovski, M, Vos, S.M, Farnung, L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosome retention during transcription elongation.
Science, 376, 2022
|
|
5OQM
 
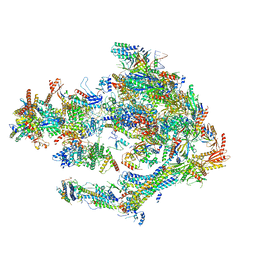 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
5OQJ
 
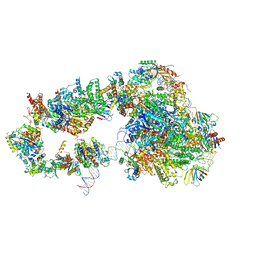 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH | | Descriptor: | DNA repair helicase RAD25, DNA repair helicase RAD3, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Henning, U, Cramer, P. | | Deposit date: | 2017-08-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
9MU4
 
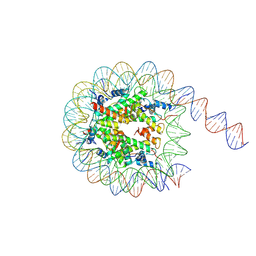 | | Structure of a native Drosophila melanogaster octameric nucleosome | | Descriptor: | DNA (164-MER), Histone H2A, Histone H2B, ... | | Authors: | Venette-Smith, N.L, Vishwakarma, R.K, Dollinger, R, Schultz, J, Venkatakrishnan, V, Babitzke, P, Anand, G, Gilmour, D.S, Armache, J.-P, Murakami, K. | | Deposit date: | 2025-01-13 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural Characterization of Native RNA Polymerase II Transcription Complexes in Drosophila melanogaster
To Be Published
|
|
9MU5
 
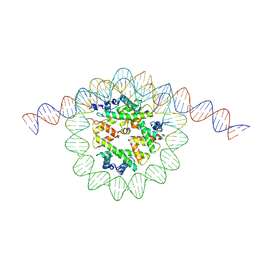 | | Structure of a native Drosophila melanogaster hexameric nucleosome | | Descriptor: | DNA (133-MER), Histone H2A, Histone H2B, ... | | Authors: | Venette-Smith, N.L, Vishwakarma, R.K, Dollinger, R, Schultz, J, Venkatakrishnan, V, Babitzke, P, Anand, G, Gilmour, D.S, Armache, J.-P, Murakami, K. | | Deposit date: | 2025-01-13 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural Characterization of Native RNA Polymerase II Transcription Complexes in Drosophila melanogaster
To Be Published
|
|
4D7X
 
 | |
