7U81
 
 | | Human DNA polymerase eta-DNA-dGMPNPP ternary mismatch complex in 0.5 mM Mn2+ for 600s | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystallo observation of three metal ion promoted DNA polymerase misincorporation.
Nat Commun, 13, 2022
|
|
5WU0
 
 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-12-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
7U7B
 
 | |
7U7S
 
 | | Human DNA polymerase eta-DNA-dGMPNPP ternary mismatch complex in 0.025 mM Mg2+ for 600s | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystallo observation of three metal ion promoted DNA polymerase misincorporation.
Nat Commun, 13, 2022
|
|
7U84
 
 | | Human DNA polymerase eta-DNA-dGMPNPP ternary mismatch complex in 6.0 mM Mn2+ for 600s | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | In crystallo observation of three metal ion promoted DNA polymerase misincorporation.
Nat Commun, 13, 2022
|
|
7U73
 
 | |
7U77
 
 | |
7U7G
 
 | |
7U7U
 
 | | Human DNA polymerase eta-DNA-dGMPNPP ternary mismatch complex in 0.1 mM Mg2+ for 600s | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In crystallo observation of three metal ion promoted DNA polymerase misincorporation.
Nat Commun, 13, 2022
|
|
1JO7
 
 | | Solution Structure of Influenza A Virus Promoter | | Descriptor: | Influenza A virus promoter RNA | | Authors: | Bae, S.-H, Cheong, H.-K, Lee, J.-H, Cheong, C, Kainosho, M, Choi, B.-S. | | Deposit date: | 2001-07-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of an influenza virus promoter and their implications for viral RNA synthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2LKP
 
 | | solution structure of apo-NmtR | | Descriptor: | HTH-type transcriptional regulator NmtR | | Authors: | Lee, C, Giedroc, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mycobacterium tuberculosis NmtR in the Apo State: Insights into Ni(II)-Mediated Allostery.
Biochemistry, 51, 2012
|
|
2LP1
 
 | | The solution NMR structure of the transmembrane C-terminal domain of the amyloid precursor protein (C99) | | Descriptor: | C99 | | Authors: | Barrett, P.J, Song, Y, Van Horn, W.D, Hustedt, E.J, Schafer, J.M, Hadziselimovic, A, Beel, A.J, Sanders, C.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol.
Science, 336, 2012
|
|
5X14
 
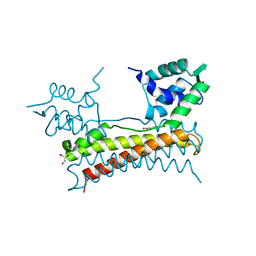 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
7K2H
 
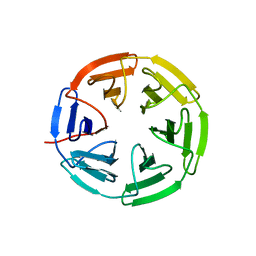 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GDPETGE] | | Descriptor: | GLY-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
1D1B
 
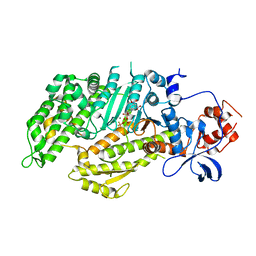 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O,P-DINITROPHENYL AMINOPROPYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, O,P-DINITROPHENYL AMINOPROPYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1JJR
 
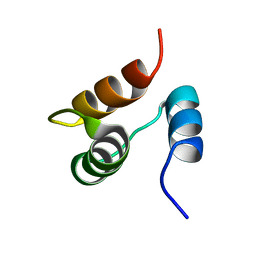 | |
7K2G
 
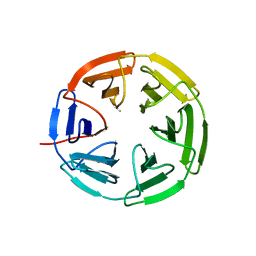 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GDEEAGE] | | Descriptor: | GLY-ASP-GLU-GLU-ALA-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
1CO9
 
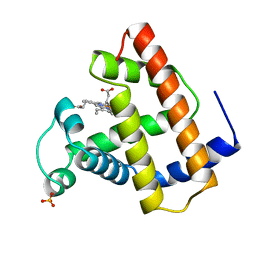 | | RECOMBINANT SPERM WHALE MYOGLOBIN L104V MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
7K2Q
 
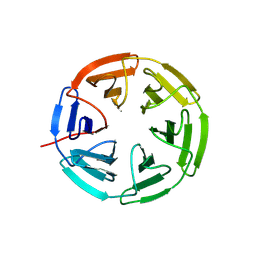 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[Ahx-DPETGE] | | Descriptor: | ACA-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
8ZJ2
 
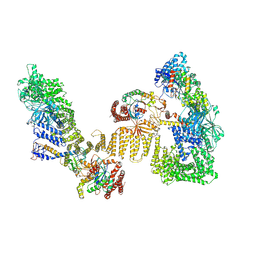 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
2C06
 
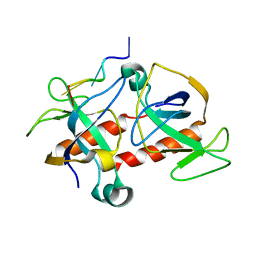 | | NMR-based model of the complex of the toxin Kid and a 5-nucleotide substrate RNA fragment (AUACA) | | Descriptor: | 5'-R(*AP*UP*AP*CP*AP)-3', KID TOXIN PROTEIN | | Authors: | Kamphuis, M.B, Bonvin, A.M.J.J, Monti, M.C, Lemonnier, M, Munoz-Gomez, A, Van Den Heuvel, R.H.H, Diaz-Orejas, R, Boelens, R. | | Deposit date: | 2005-08-25 | | Release date: | 2006-02-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Model for RNA Binding and the Catalytic Site of the Rnase Kid of the Bacterial Pard Toxin-Antitoxin System.
J.Mol.Biol., 357, 2006
|
|
2WVT
 
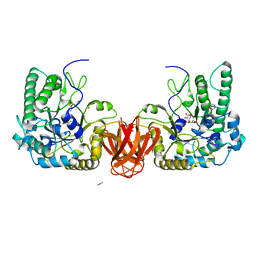 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron in complex with a novel iminosugar fucosidase inhibitor | | Descriptor: | (2S,3R,5R,6S)-3,4,5-TRIHYDROXY-2,6-BIS(HYDROXYMETHYL)PIPERIDINIUM, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach.
J.Am.Chem.Soc., 132, 2010
|
|
5MFC
 
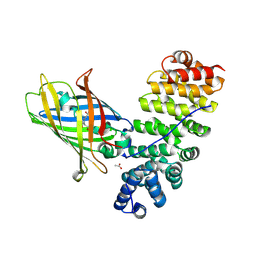 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
7JYZ
 
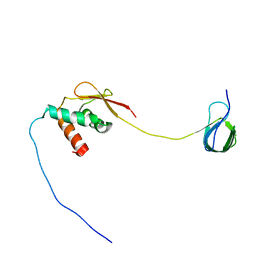 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
3F92
 
 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington Interacting Protein 2) M172A mutant crystallized at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Wilson, R.C, Hughes, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-11-13 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
