4COQ
 
 | | The complex of alpha-Carbonic anhydrase from Thermovibrio ammonificans with inhibitor sulfanilamide. | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CARBONATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J.A. | | Deposit date: | 2014-01-30 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZCL
 
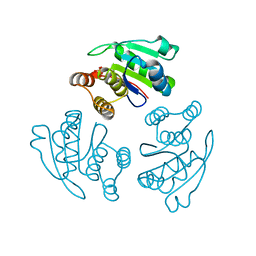 | | prl-1 c104s mutant in complex with sulfate | | Descriptor: | SULFATE ION, protein tyrosine phosphatase 4a1 | | Authors: | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
2GQ5
 
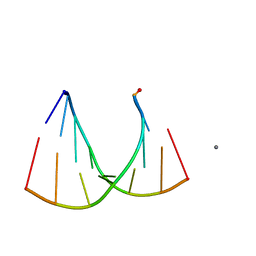 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2Z20
 
 | | Crystal structure of LL-Diaminopimelate Aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Cherney, M.M, van Belkum, M.J, Marcus, S.L, Flegel, M.D, Clay, M.D, Deyholos, M.K, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia
J.Mol.Biol., 371, 2007
|
|
1N8T
 
 | |
4CKQ
 
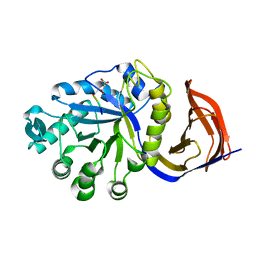 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum | | Descriptor: | 4 HISTIDINES FROM PROTEOLYSED HIS-TAG, CARBOHYDRATE BINDING FAMILY 6, MALONIC ACID | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-01-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1NA2
 
 | |
1EZN
 
 | | SOLUTION STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA THREE-WAY JUNCTION | | Authors: | van Buuren, B.N.M, Overmars, F.J, Ippel, J.H, Altona, C, Wijmenga, S.S. | | Deposit date: | 2000-05-11 | | Release date: | 2001-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA three-way junction containing two unpaired thymidine bases. Identification of sequence features that decide conformer selection.
J.Mol.Biol., 304, 2000
|
|
1NGJ
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1PFG
 
 | | Strategy to design inhibitors: Structure of a complex of Proteinase K with a designed octapeptide inhibitor N-Ac-Pro-Ala-Pro-Phe-DAla-Ala-Ala-Ala-NH2 at 2.5A resolution | | Descriptor: | N-Ac-PAPFAAAA-NH2, Proteinase K | | Authors: | Saxena, A.K, Singh, T.P, Peters, K, Fittkau, S, Betzel, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Strategy to design peptide inhibitors: structure of a complex of proteinase K with a designed octapeptide inhibitor N-Ac-Pro-Ala-Pro-Phe-DAla-Ala-Ala-Ala-NH2 at 2.5 A resolution.
Protein Sci., 5, 1996
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
2GI0
 
 | |
5DBA
 
 | |
3BIQ
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16, GLYCEROL | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
4GNE
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
1MBE
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
1MBG
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
3BIT
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FACT complex subunit SPT16, ... | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
2ANB
 
 | | Crystal Structure Of Oligomeric E.coli Guanylate Kinase In Complex With GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, SULFATE ION | | Authors: | Hible, G, Renault, L, Schaeffer, F, Christova, P, Radulescu, A.Z, Evrin, C, Gilles, A.M, Cherfils, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase
J.Mol.Biol., 352, 2005
|
|
1MBK
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
5DHV
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHJ
 
 | | PIM1 in complex with Cpd4 (3-methyl-5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridine) | | Descriptor: | 3-methyl-5-(pyridin-3-yl)-2H-pyrazolo[3,4-c]pyridine, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1IQT
 
 | | Solution structure of the C-terminal RNA-binding domain of heterogeneous nuclear ribonucleoprotein D0 (AUF1) | | Descriptor: | heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Katahira, M, Miyanoiri, Y, Enokizono, Y, Matsuda, G, Nagata, T, Ishikawa, F, Uesugi, S. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA.
J.Mol.Biol., 311, 2001
|
|
