2QIC
 
 | | Crystal Structure of the ING1 PHD Finger in complex with a Histone H3K4ME3 peptide | | Descriptor: | H3K4ME3 PEPTIDE, Inhibitor of growth protein 1, ZINC ION | | Authors: | Pena, P.V, Champagne, K, Zhao, R, Kutateladze, T.G. | | Deposit date: | 2007-07-03 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor.
J.Mol.Biol., 380, 2008
|
|
6KC0
 
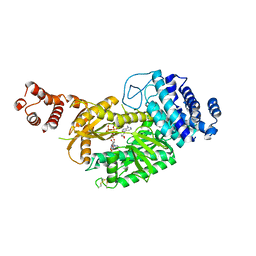 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
9CER
 
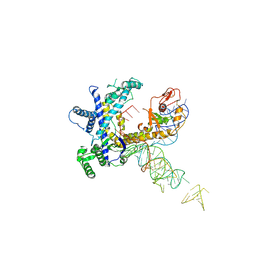 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
8DFU
 
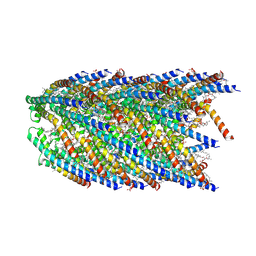 | | Cryo-EM structure of conjugation pili from Aeropyrum pernix | | Descriptor: | (2S)-3-{[(3R,7S,11S,15S)-3,7,11,15,19-pentamethylicosyl]oxy}-2-{[(2R,6S,10S,14R)-2,6,10,14,18-pentamethylnonadecyl]oxy}propyl dihydrogen phosphate, Pilin protein | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
8DFT
 
 | | Cryo-EM structure of conjugative pili from Pyrobaculum calidifontis | | Descriptor: | Pilin protein, [(2~{S},7~{S},11~{S},15~{S},19~{R},22~{R},26~{S},30~{R},34~{R},38~{S},43~{S},47~{S},51~{S},55~{R},58~{R},62~{S},66~{R},70~{R})-38-(hydroxymethyl)-7,11,15,19,22,26,30,34,43,47,51,55,58,62,66,70-hexadecamethyl-1,4,37,40-tetraoxacyclodoheptacont-2-yl]methanol | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
6TUN
 
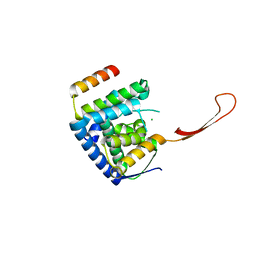 | | Helicase domain complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, CHLORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPD | | Authors: | Sauer, F, Kisker, C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair.
Nat Commun, 11, 2020
|
|
9GD3
 
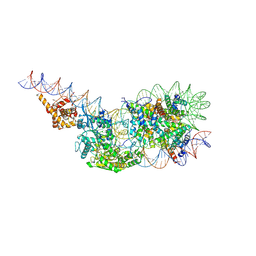 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
7P3F
 
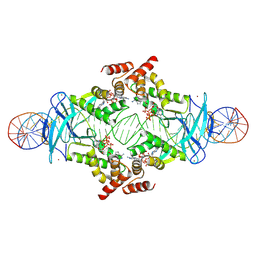 | |
4Y52
 
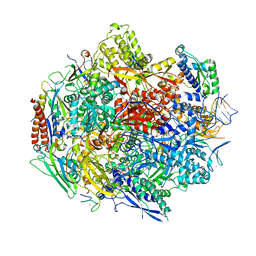 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
6MZL
 
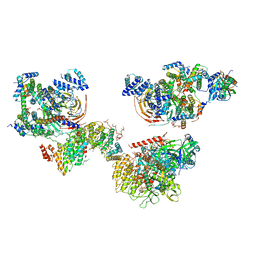 | | Human TFIID canonical state | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
6TUO
 
 | |
7OMK
 
 | | The NMR structure of the Zf-GRF domains from the mouse Endonuclease VIII-LIKE 3 (mNEIL3) | | Descriptor: | Endonuclease 8-like 3 | | Authors: | Dinesh, D.C, Huskova, A, Srb, P, Veverka, V, Silhan, J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Model of abasic site DNA cross-link repair; from the architecture of NEIL3 DNA binding domains to the X-structure model.
Nucleic Acids Res., 50, 2022
|
|
8ZVI
 
 | | Structure of the bacteriophage T5 capsid | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Peng, Y, Liu, H.R. | | Deposit date: | 2024-06-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
7QV3
 
 | |
6MZD
 
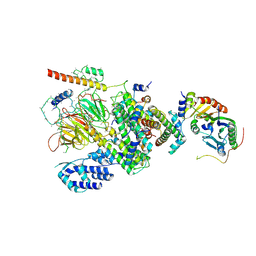 | | Human TFIID Lobe A canonical | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
4WVT
 
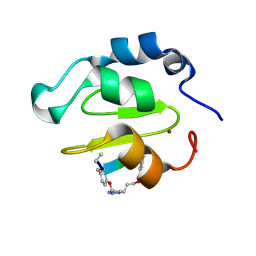 | | Crystal structure of XIAP-BIR2 domain complexed with ligand bound | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
4WVU
 
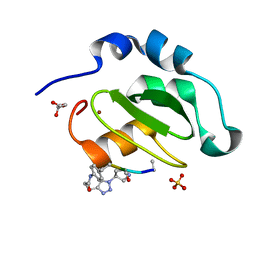 | | CRYSTAL STRUCTURE OF XIAP-BIR2 DOMAIN COMPLEXED WITH LIGAND BOUND | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
6MZC
 
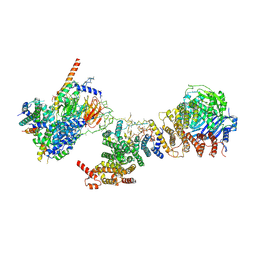 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
9ILP
 
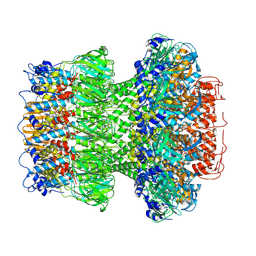 | | Structure of the bacteriophage T5 portal complex | | Descriptor: | Head completion protein, Portal protein pb7 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9ILV
 
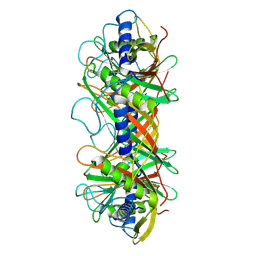 | | Structure of the bacteriophage T5 connector complex | | Descriptor: | Tail tube terminator protein p142 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9IOZ
 
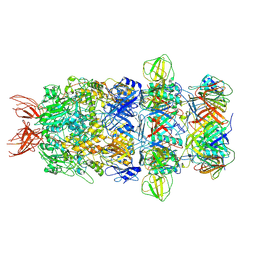 | | Structure of the bacteriophage T5 tail tip complex | | Descriptor: | Baseplate hub protein pb3, Baseplate tube protein p140, Distal tail protein pb9 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9INY
 
 | | Structure of bacteriophage T5 tail tube | | Descriptor: | Tail tube protein pb6 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9IMH
 
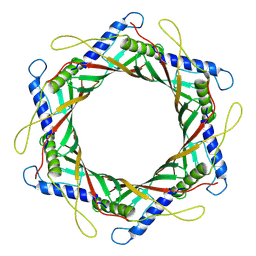 | |
9IMV
 
 | |
