7D27
 
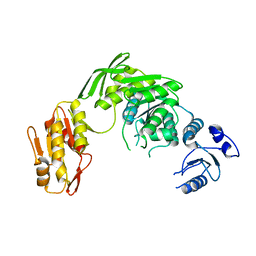 | |
4BJR
 
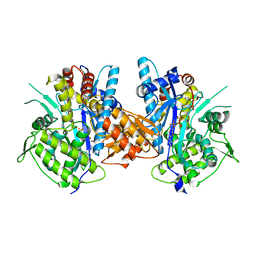 | | Crystal structure of the complex between Prokaryotic Ubiquitin-like Protein Pup and its Ligase PafA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE, ... | | Authors: | Barandun, J, Delley, C.L, Ban, N, Weber-Ban, E. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Complex between Prokaryotic Ubiquitin-Like Protein Pup and its Ligase Pafa.
J.Am.Chem.Soc., 135, 2013
|
|
1VLB
 
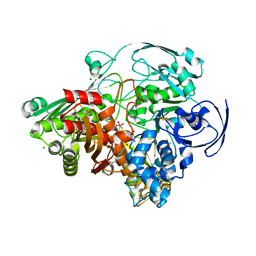 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
8R53
 
 | | The complex of Glycogen Phosphorylase with (-)-Epigallocatechin-3-gallate (EGCG) and glucose. | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-11-15 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
8QMU
 
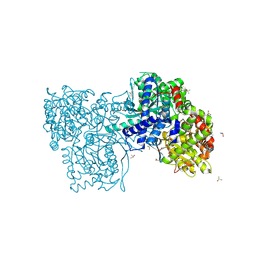 | | The complex of Glycogen Phosphorylase with (-)-Epigallocatechin-3-gallate (EGCG). | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
1IQA
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MOUSE RANK LIGAND | | Descriptor: | RECEPTOR ACTIVATOR OF NUCLEAR FACTOR KAPPA B LIGAND | | Authors: | Ito, S, Wakabayashi, K, Ubukata, O, Hayashi, S, Okada, F, Hata, T. | | Deposit date: | 2001-07-11 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the extracellular domain of mouse RANK ligand at 2.2-A resolution.
J.Biol.Chem., 277, 2002
|
|
8XA7
 
 | | Cryo-EM structure of Bacillus subtilis RNAP,sigA and SPO1 gp33 complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, M, Jin, Q, Yuan, S, Liu, B. | | Deposit date: | 2023-12-02 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A phage encoded sigA-dependent transcription inhibitor also attacks host HelD-mediated defence system
To Be Published
|
|
7SNU
 
 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | Descriptor: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
1H0H
 
 | | Tungsten containing Formate Dehydrogenase from Desulfovibrio Gigas | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Raaijmakers, H.C.A. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gene Sequence and the 1.8 A Crystal Structure of the Tungsten-Containing Formate Dehydrogenase from Desulfovibrio Gigas
Structure, 10, 2002
|
|
1SIJ
 
 | | Crystal structure of the Aldehyde Dehydrogenase (a.k.a. AOR or MOP) of Desulfovibrio gigas covalently bound to [AsO3]- | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ARSENITE, Aldehyde oxidoreductase, ... | | Authors: | Boer, D.R, Thapper, A, Brondino, C.D, Romao, M.J, Moura, J.J.G. | | Deposit date: | 2004-03-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure and EPR Spectra of "Arsenite-Inhibited" Desulfovibriogigas Aldehyde Dehydrogenase: A Member of the Xanthine Oxidase Family
J.Am.Chem.Soc., 126, 2004
|
|
8R52
 
 | | The complex of Glycogen Phosphorylase with epigallocatechin (EGC). | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-11-15 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
8R6V
 
 | | The complex of glycogen phosphorylase with EGCG (epigallocatechin gallate) and caffeine. | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-11-23 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
6GCJ
 
 | | Solution structure of the RodA hydrophobin from Aspergillus fumigatus | | Descriptor: | Hydrophobin | | Authors: | Pille, A, Kwan, A, Aimanianda, V, Latge, J.-P, Sunde, M, Guijarro, J.I. | | Deposit date: | 2018-04-18 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Assembly and disassembly of Aspergillus fumigatus conidial rodlets
Cell Surf, 2019
|
|
6ZPS
 
 | |
6ZPX
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPW
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPV
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8EXD
 
 | |
7RHV
 
 | | Aspergillus fumigatus Enolase | | Descriptor: | Enolase, MAGNESIUM ION | | Authors: | Nguyen, S, Bruning, J.B. | | Deposit date: | 2021-07-19 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural model of the human plasminogen and Aspergillus fumigatus enolase complex.
Proteins, 90, 2022
|
|
7RI0
 
 | |
7RHW
 
 | |
1IMO
 
 | |
7YTV
 
 | |
8JEA
 
 | | Crystal structure of CGL1 from Crassostrea gigas, mannotriose-bound form (CGL1/Man(alpha)1-2Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
8JE9
 
 | | Crystal structure of CGL1 from Crassostrea gigas, mannobiose-bound form (CGL1/Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, Natterin-3, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
