8IEO
 
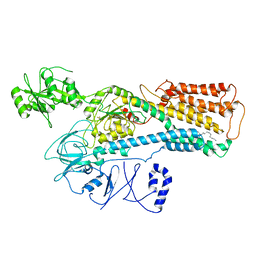 | | Cryo-EM structure of ATP13A2 in the nominal E1P state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8I2S
 
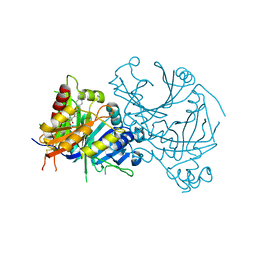 | | Crystal structure of AtHPPD-Y18979 complex | | Descriptor: | 1,5-dimethyl-3-(naphthalen-2-ylmethyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystal structure of AtHPPD-Y18979 complex
To Be Published
|
|
8HYM
 
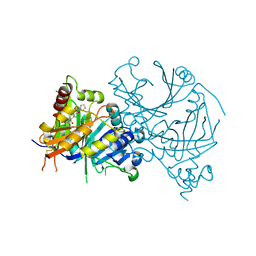 | | Crystal structure of AtHPPD-Y18030 complex | | Descriptor: | 3-cyclopentyl-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of AtHPPD-Y18030 complex
To Be Published
|
|
8I15
 
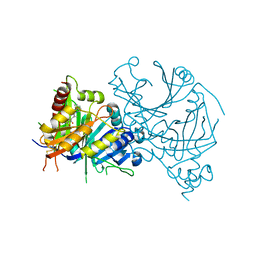 | | Crystal structure of AtHPPD-YH20335 complex | | Descriptor: | 1,4-dimethyl-5-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)benzimidazol-2-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-12 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of AtHPPD-YH20335 complex
To Be Published
|
|
2RQE
 
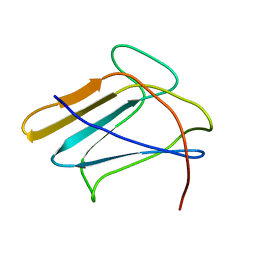 | | Solution structure of the silkworm bGRP/GNBP3 N-terminal domain reveals the mechanism for b-1,3-glucan specific recognition | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Takahasi, K, Ochiai, M, Horiuchi, M, Kumeta, H, Ogura, K, Ashida, M, Inagaki, F. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8I2U
 
 | | Crystal structure of AtHPPD-YH20282 complex | | Descriptor: | 2-[2-chloranyl-4-methylsulfonyl-3-(2-trimethylsilylethoxy)phenyl]carbonyl-3-oxidanyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of AtHPPD-YH20282 complex
To Be Published
|
|
8IEL
 
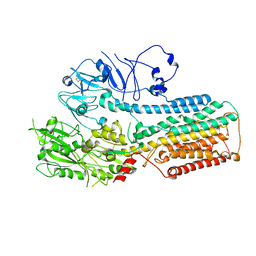 | |
8IB8
 
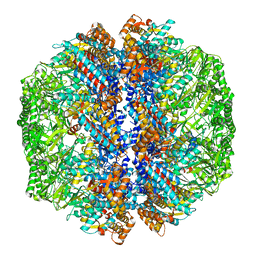 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
4UY5
 
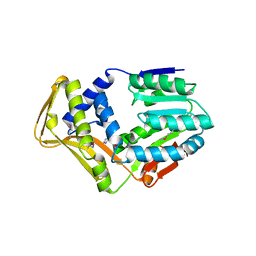 | |
8HX2
 
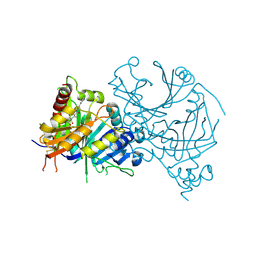 | | Crystal structure of AtHPPD-Y18405 complex | | Descriptor: | 3-[2-(3-chlorophenyl)ethyl]-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexa-1,3-dien-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-03 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structure of AtHPPD-Y18405 complex
To Be Published
|
|
8HZ9
 
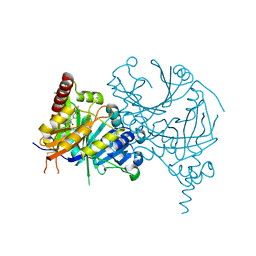 | | Crystal structure of AtHPPD-Y181136 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-3-propan-2-yl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of AtHPPD-Y181136 complex
To Be Published
|
|
2RS2
 
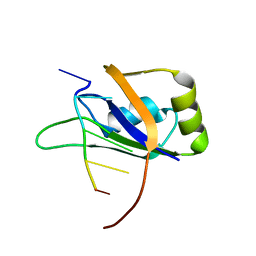 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
4PGB
 
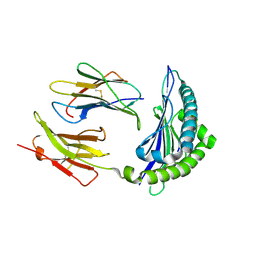 | | MHC Class I in complex with modified Sendai virus nucleoprotein peptide FAPGNWPAL | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Celie, P.H.N, Joosten, R.P, Perrakis, A, Neefjes, J. | | Deposit date: | 2014-05-01 | | Release date: | 2015-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The first step of peptide selection in antigen presentation by MHC class I molecules.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8HZ6
 
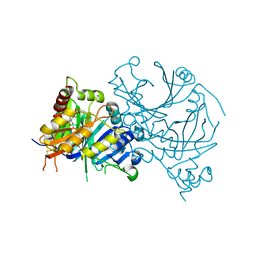 | | Crystal structure of AtHPPD-QRY2089 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-prop-2-ynyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Crystal structure of AtHPPD-QRY2089 complex
To Be Published
|
|
4PGP
 
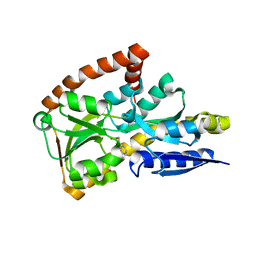 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND 3-INDOLE ACETIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
8IDU
 
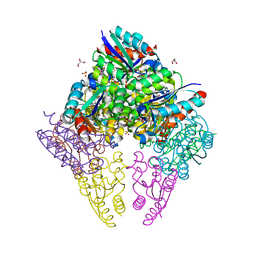 | | Crystal structure of substrate bound-form dehydroquinate dehydratase from Corynebacterium glutamicum | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL, ... | | Authors: | Lee, C.H, Kim, S, Kim, K.-J. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Analysis of 3-Dehydroquinate Dehydratase from Corynebacterium glutamicum .
J Microbiol Biotechnol., 33, 2023
|
|
8IEK
 
 | |
8IGI
 
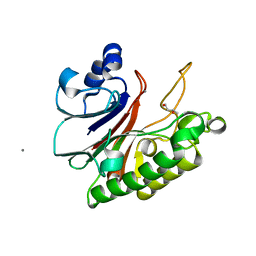 | | Crystal structure of HP1526 (XthA)- a base excision DNA repair protein in Helicobacter pylori | | Descriptor: | 1,3-BUTANEDIOL, Exodeoxyribonuclease (LexA), MANGANESE (II) ION | | Authors: | Dinh, T.T, Dao, O, Lee, K.H. | | Deposit date: | 2023-02-20 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease XthA (HP1526 protein) from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 663, 2023
|
|
2THF
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225F MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, THROMBIN HEAVY CHAIN, ... | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2TMV
 
 | |
8HWD
 
 | | Cryo-EM Structure of D5 ADP form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primase D5 | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2TPK
 
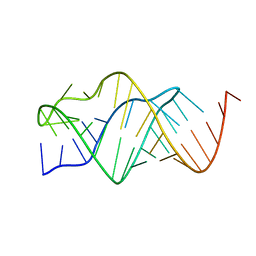 | |
8HYN
 
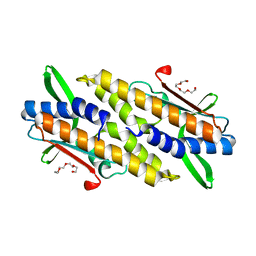 | | Bacterial STING from Riemerella anatipestifer | | Descriptor: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8IDR
 
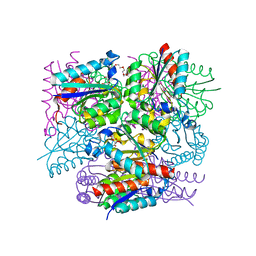 | |
4W82
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase | | Descriptor: | CHLORIDE ION, Fatty acid synthase, MAGNESIUM ION, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
