8WFW
 
 | |
8WFV
 
 | |
8WFU
 
 | |
8WFT
 
 | |
8WFR
 
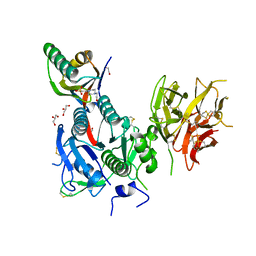 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
8WF5
 
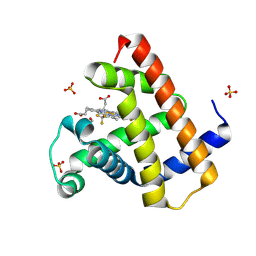 | | Horse heart myoglobin reconstituted with an iron complex of porphyrin bearing two CF3 groups (rMb(FePor(CF3)2)) | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN(CF3)2 CONTAINING FE, ... | | Authors: | Kagawa, Y, Oohora, K, Himiyama, T, Suzuki, A, Hayashi, T. | | Deposit date: | 2023-09-19 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Redox Engineering of Myoglobin by Cofactor Substitution to Enhance Cyclopropanation Reactivity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8WEJ
 
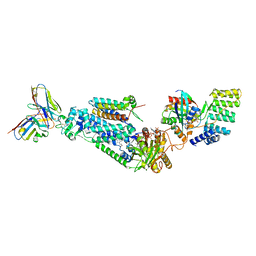 | |
8WEA
 
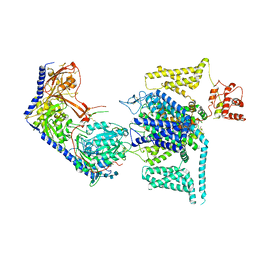 | | Human L-type voltage-gated calcium channel Cav1.2 (Class II) in the presence of pinaverium at 3.2 Angstrom resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE9
 
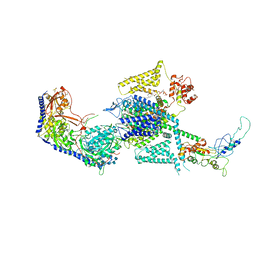 | | Human L-type voltage-gated calcium channel Cav1.2 (Class I) in the presence of pinaverium at 3.0 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE8
 
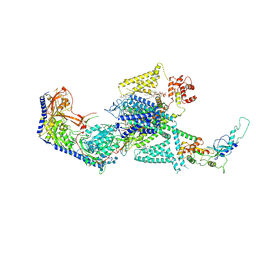 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine, amlodipine and pinaverium at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE7
 
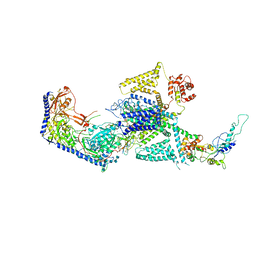 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine at 3.2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE6
 
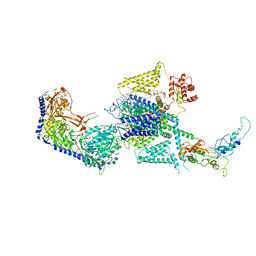 | | Human L-type voltage-gated calcium channel Cav1.2 at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDT
 
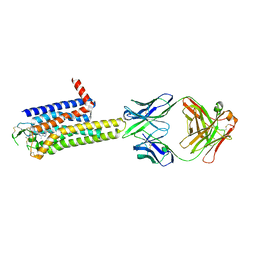 | | Crystal structure of the human adenosine A2A receptor in complex with photoresponsive ligand photoNECA(blue) | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-2-((E)-phenyldiazenyl)-9H-purin-9-yl)-N-ethyl-3,4-dihydroxytetrahydrofuran-2-carboxamide, Adenosine receptor A2a, Antibody Fab fragment heavy chain, ... | | Authors: | Araya, T, Asada, H, Iwata, S, Im, D.H. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure reveals the binding mode and selectivity of a photoswitchable ligand for the adenosine A 2A receptor.
Biochem.Biophys.Res.Commun., 695, 2023
|
|
8WDO
 
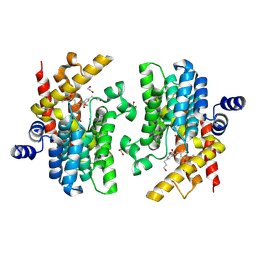 | | Crystal structure of PDE4D complexed with DCN | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-(3,4-dimethoxyphenyl)-3-(hydroxymethyl)-7-methoxy-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
8WDN
 
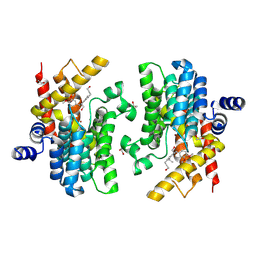 | | Crystal structure of PDE4D complexed with 7b-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-7-ethoxy-2-(3-ethoxy-4-methoxy-phenyl)-3-(hydroxymethyl)-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
8WDG
 
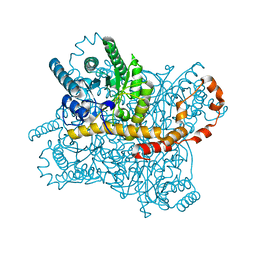 | |
8WD8
 
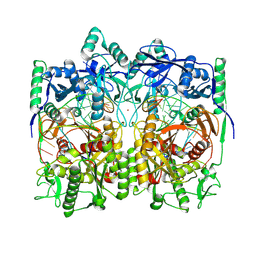 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
8WD5
 
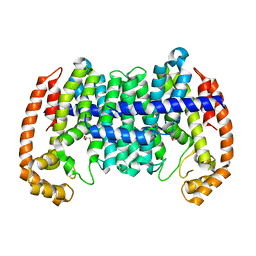 | |
8WD3
 
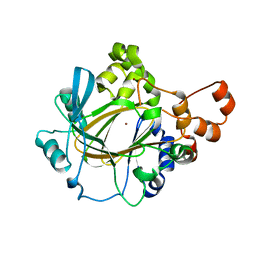 | | The Crystal Structure of JMJD2A(M1-L359) from Biortus. | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2023-09-14 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of JMJD2A(M1-L359) from Biortus.
To Be Published
|
|
8WD0
 
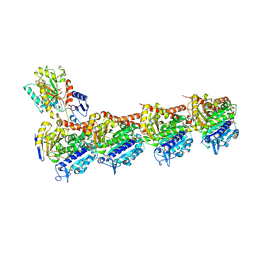 | | Crystal structure of T2R-TTL-Erianin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-[2-(3,4,5-trimethoxyphenyl)ethyl]phenol, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cytotoxic natural compound erianin binds to colchicine site of beta-tubulin and overcomes taxane resistance
Bioorg.Chem., 150, 2024
|
|
8WCT
 
 | |
8WCS
 
 | | Cryo-EM structure of Cas13h1-crRNA binary complex | | Descriptor: | 66-nt crRNA, Cas13h1, MAGNESIUM ION | | Authors: | Zhang, C. | | Deposit date: | 2023-09-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism for target RNA recognition and cleavage of Cas13h.
Nucleic Acids Res., 52, 2024
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
